Phylogenetic and ecological constraints on fighting
advertisement

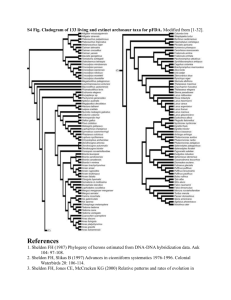
Supplementary information for Nature’s web site Figure A. Molecular phylogenies of 15 species of fig wasps, estimated using nucleotide sequences 816 base pair long collected from the 3’ end of the cytochrome oxidase subunit I gene (COI)1. Data was deposited in GenBank (Accession numbers AF302052 to AF302067). Analyses were carried out with version 4.0b1 of PAUP*, written by David L. Swofford. Maximum likelihood methods were used to reconstruct the phylogeny. The general reversible model with rate heterogeneity was used2, and the parameters of the model were estimated from the data (proportion of sites assumed to be invariable = 0.284265; rates (for variable sites) assumed to follow a gamma distribution with shape parameter = 0.865608). The tree topology was estimated using an heuristic algorithm with branch swapping (tree bisection-reconnection). The heuristic search was repeated 3 times and the consensus tree from the best 3 trees was used for generating the final phylogeny. Here we show the topology of two of the 3 final trees (-log (L) = 5679.00011). Numbers above branches are the bootstrap values (>50%; 500 replications) for the same nodes of the neighbour joining (NJ) tree reconstructed using Tamura-Nei distances3. The NJ tree has an almost identical topology as the ML trees, the only major difference being the well supported monophyly of genus Ceratosolen (85% bootstrap support). The monophyly of Ceratosolen is highly supported by additional sequence data1 and by morphology. The sequences of C. solmsi and C. constrictus are part of a lineage of fig pollinators in which there is a significant increase in substitution rate in COI and COII1. Figure B. Consensus molecular based phylogeny of the species of fig wasp for which injury levels were scored. The species scored for male injury level that were not present in the molecular phylogeny (Figure A) were added to the nodes leading to species of the same genus or subfamily. The topology of this phylogeny agrees completely with other molecular data1,4,5. REFERENCES 1. Machado, C. A. Molecular Natural History of Fig Wasps 1-187 (University of California, Irvine, CA, 1998). 2. Yang, Z. Estimating the pattern of nucleotide substitution J. Molec. Evol. 39, 105-111 (1994). 3. Tamura, K. & Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10, 512-526 (1993). 4. Machado, C. A., Herre, E. A., McCafferty, S. & Bermingham, E. Molecular phylogenies of fig pollinating and non-pollinating wasps and the implications for the origin and evolution of the fig-fig wasp mutualism J. Biogeogr. 23, 531-542 (1996). 5. Rasplus, J.-Y., Kerdelhue, C., Le Clainche, I. & Mondor, G. Molecular phylogeny of fig wasps. Agaonidae are not monophyletic. Comptes Rendus de l’Académie des Sciences de Paris, Sciences de la vie/Life Sciences 321, 517-527 (1998). 1 A. Blastophaga nipponica 98 Eupristina verticillata Ceratosolen galili Ceratosolen solmsi 100 Ceratosolen constrictus Eujacobsonia sp 100 Apocrypta sp Sycoryctes sp 88 Sycoscapter sp Philotrypesis sp Heterandrium sp Eukoebelea sp 97 Apocryptophagus sp 99 Idarnes sp6 99 Idarnes sp2 0.05 substitutions/site Blastophaga nipponica 98 Eupristina verticillata Ceratosolen galili Ceratosolen solmsi 100 Ceratosolen constrictus Eujacobsonia sp Philotrypesis sp 100 Apocrypta sp 88 Sycoryctes sp Sycoscapter sp Heterandrium sp 97 Eukoebelea sp Apocryptophagus sp 99 99 Idarnes sp6 Idarnes sp2 0.05 substitutions/site 2 B. Blastophaga sp. Subfamily Deilagaon annulatae Eupristina adempta Ceratosolen sp. Agaoninae (pollinators) Ceratosolen solmsi Ceratosolen constrictus Apocrypta bakeri Apocrypta varicolor Sycorictes sp1 Sycorictes sp2 Sycoscapter sp1 Sycoryctinae Sycoscapter sp2 Sycoscapter sp3 Arachonia sp. Eujacobsonia genalis Otites ellinae Philotrypesis spinipes Philotrypesis pilosa Philotrypesis sp1 Sycoryctinae Philotrypesis sp2 Philotrypesis sp3 Lypothymus sundaicus Otitesellinae sp. Otites ellinae Epychrisomallinae sp. Epychrysomallinae Eukoebelea sp1 Eukoebelea sp2 3 Sycophaginae