RNA
advertisement
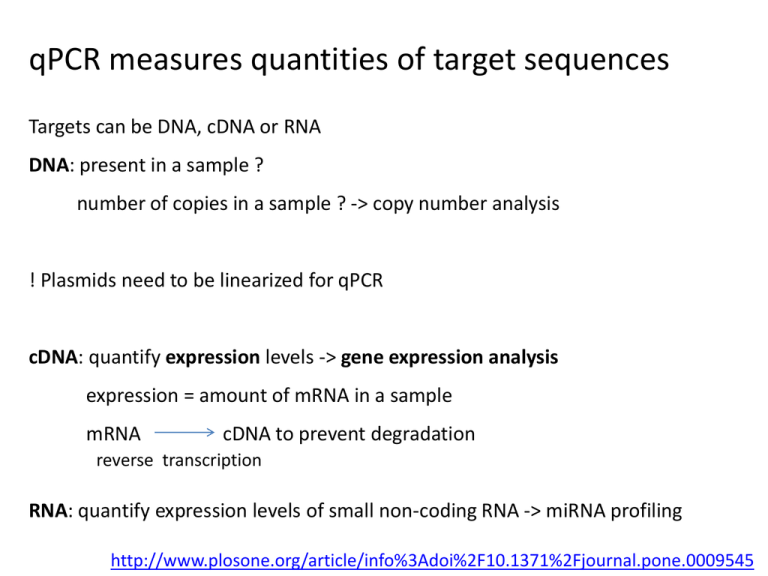
qPCR measures quantities of target sequences Targets can be DNA, cDNA or RNA DNA: present in a sample ? number of copies in a sample ? -> copy number analysis ! Plasmids need to be linearized for qPCR cDNA: quantify expression levels -> gene expression analysis expression = amount of mRNA in a sample mRNA cDNA to prevent degradation reverse transcription RNA: quantify expression levels of small non-coding RNA -> miRNA profiling http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0009545 qPCR protocol involves many steps Growing the samples RNA extraction DNase treatment cDNA synthesis Primer design qPCR Starting with good samples: RNA extraction mRNA or miRNA extraction ? -> different kits recommended: use a kit that can extract both e.g. Qiagen miRNeasy use same samples for mRNA and miRNA profiling Check RNA quality - Integrity: RNA breaks down easily - Purity: RNA can contain inhibitors of qPCR reaction ! Don’t aim for high quality, aim for equal quality between samples ! Checking RNA integrity by microfluidic electrophoresis e.g. Agilent Bioanalyzer, Bio-Rad Experion Evaluates integrity of 18S and 28S rRNA You look at rRNA while you’re interested in mRNA If you see these peaks there’s no degradation Checking RNA integrity by 5’-3’ mRNA ratio Perform reverse transcription on RNA Use qPCR to measure HPRT1 in cDNA using two primer sets HPRT1 5’ Cq 3’ Cq 5’ No degradation: Cq 5’ = Cq 3’ Degradation: Cq 5’ <<< Cq 3’ HPRT1 is a reference gene with stable, low expression levels HPRT1 is expressed in all organisms AAAAAAA 3’ Inhibitors distort the qPCR reaction No inhibitors Inhibitors: shift Cq to right e.g. phenol, ethanol... Checking RNA purity by SPUD assay SPUD = potato gene with no homology to any known sequence Add equal amounts SPUD to each RNA sample Create controls: positive: SPUD + heparin (a known inhibitor) negative: SPUD + water Perform qPCR to measure SPUD SPUD + water SPUD + heparin SPUD + RNA1 SPUD + RNA2 SPUD + RNA3 Cq=22 Cq=27 Cq=22 Cq=26 Cq=22 Clean samples have same Cq as water Samples with inhibitors have higher Cq ΔCq > 1: presence of inhibitors Starting with good samples: DNase treatment To remove genomic DNA contamination from mRNA samples Recommended but never 100% efficient Kits: Qiagen RNase-Free DNase Set for DNase digestion during RNA purification Solutions: • Intron or exon-exon junction spanning primers: not always feasible gDNA will generate a longer or no PCR product • No RT control to detect gDNA contamination Starting with good samples: cDNA synthesis To transform RNA into more stable cDNA Prepare all cDNAs that you are going to use in a single batch Kits: Biogazelle uses BioRad iScript Advanced Primers: Biogazelle uses a mix of random and oligodT primers Validation: RNA RNA 1/4 RNA 1/16 RNA 1/64 RNA 1/256 cDNA cDNA cDNA cDNA cDNA Cq=18 Cq=20 Cq=22 Cq=24 Cq=26 Create RNA dilution series Synthesize cDNA qPCR Cqs follow dilution series: good cDNA synthesis cDNA synthesis + DNase treatment directly on cells: Ambion Cells-to-CT kit Different ways to measure amount of PCR product Fluorescent dyes: e.g. Sybr Green, EvaGreen... Fluorescent dye binds to ds DNA Denaturation: dye is released and fluorescence reduces Polymerization: primers anneal and ds PCR product is formed Dye binds to ds product and fluorescence increases Fluorophore-containing DNA probes: e.g. Taqman... Plot of the fluorescent signal (Rn) during PCR Rn Signal of unbound dye Cycle Threshold for the transition of baseline to exponential phase ∆Rn = Rn -baseline ABI instruments use different thresholds for different plates ! Not Ok: Use the same threshold on every plate ! qPCR generates Relative Quantities (RQ) more DNA/RNA less DNA/RNA ∆Cq = difference between 2 Cq’s 0 1 2 3 4 10 11... Detection of abnormal amplification: visual inspection of the curves Inspection of melting curves Inspection of melting curves qPCR primer design guidelines regarding PCR products degraded material: 50-80 bp good material: 80-150 bp avoid repeats and domains (high conservation: no specific primers) take into account splice variants and SNPs qPCR primer design software Primer Design Primer3Plus: http://primer3plus.com/cgi-bin/dev/primer3plus.cgi PrimerQuest: https://eu.idtdna.com/PrimerQuest/Home/Index Primer-BLAST: http://blast.ncbi.nlm.nih.gov/ Checking primer specificity BLAST: http://blast.ncbi.nlm.nih.gov/ BiSearch: http://bisearch.enzim.hu/ Checking secondary structures in the PCR product UNAfold: http://mfold.rna.albany.edu/ Checking location of SNPs UCSC in silico PCR + SNPs track: http://genome.ucsc.edu/cgi-bin/hgPcr UNAfold secondary structure prediction of PCR product Secondary structure overlapping primer annealing site Do not use these primers ! qPCR primer design guidelines regarding primers intron or exon-exon junction spanning length: 9-30 bp (ideally: 20 bp) melting temperature Tm: 58-60 °C (ideally: 59°C) maximum Tm difference between primers: 2°C GC content: 30-80% (ideally: 50%) 5 nucleotides at 3’ end < 3 G or C avoid runs > 3 identical nucleotides Same primers ordered from different vendors will give different results -> variation in amount of inhibitors in the primers Recommended: IDT primers Checking primer / probe specificity Primer specificity Always: inspection of melting curves First time use: gel analysis Probe specificity Negative control (sample in which gene is not expressed) Take home message: overview of recommendations Different location pre- and post PCR 96 well versus 384 well cheaper more data easier to pipet smaller volumes Manual versus robotic pipetting 96 well 384 well multichannel pipets: calibrate regularly ! Never use volumes < 1 µl (pipetting errors too high) Check quality of pipets: pilot experiment with same sample-target in all wells -> differences in Cq should be < 0.2 Take home message: overview of recommendations Replicates Choose biological replicates over technical replicates 4 biological replicates Controls Negative controls: no template, no RT, biological control Positive control: biological control Reference genes Minimum 3 Validate primers, kits and reference genes before the experiment ! Take home message: overview of recommendations Sample maximization Put all samples of the same gene on the same plate No need to repeat reference genes on each plate When you want to compare one gene over different conditions <-> when you want to compare genes: gene maximization If samples of same gene are to be spread over different plates: use IRCs