Proteins - Postezy
advertisement
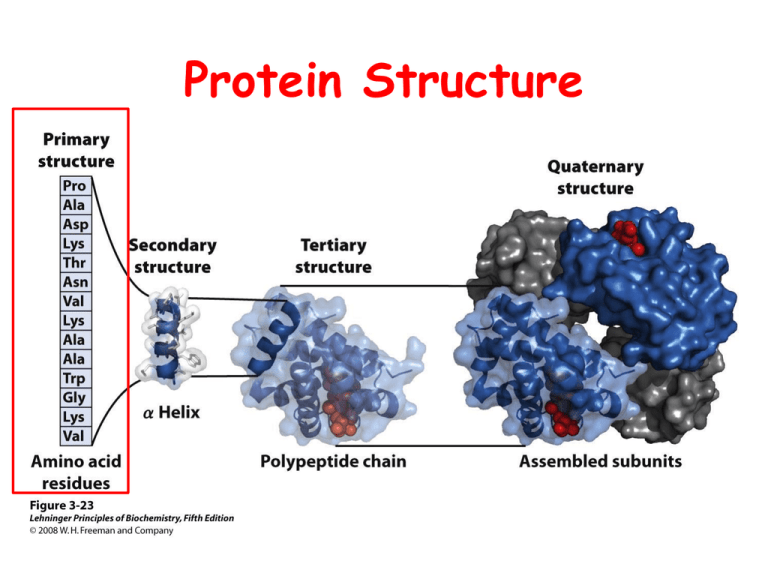
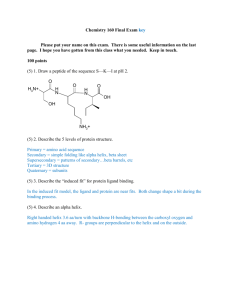
Protein Structure Protein Structure I Primary Structure Primary Structure Insulin Bovine: Insulin Figure 5-1 Human: ProInsulin Signal sequence Chain B MALWMRLLPLLALLALWGPDPAAAFVNQHLCGSHLVEALYLV C Peptide CGERGFFYTPKTRREAEDLQVGQVELGGGPGAGSLQPLALEG Chain A SLQKRGIVEQCCTSICSLYQLENYCN Primary Structure Insulin Bovine: Insulin Figure 5-1 Human: ProInsulin Signal sequence Chain B MALWMRLLPLLALLALWGPDPAAAFVNQHLCGSHLVEALYLV C Peptide CGERGFFYTPKTRREAEDLQVGQVELGGGPGAGSLQPLALEG Chain A SLQKRGIVEQCCTSICSLYQLENYCN Value of Primary Structure Information • Primary sequence information is – prerequisite for determining three-dimensional structure – essential in understanding molecular mechanism of action • Sequence comparisons among analogous proteins – provide insights into protein function – reveal evolutionary relationships • Sequence of proteins whose mutations result in inherited diseases – assist in development of diagnostic tests – assist in development of effective therapies Primary Structure Determination Strategy • Purification of protein to homogeneity • Prepare protein for sequencing • Sequence polypeptide chains • Organize completed structure Alternative: Nucleic Acid Sequencing Sequencing Strategy Summary Figure 5-12 Sequencing Strategy I Figure 5-12 Sequencing Strategy II Figure 5-12 Sequencing Strategy III Figure 5-12 Purification of Protein to Homogeneity Prepare Protein for Sequencing • End Group Analysis: How many different subunits • Cleavage of disulfide bonds • Separation and purification of the polypeptide chains • Amino acid composition End Group Analysis (How Many Different Subunits?) N-Terminal Identification Sanger’s Reagent H+ + HF NO2 R + O2N F H3N NO2 O CH C O2N DNFB (2,4 Dinitrofluorobenzene) R NH CH C H+ NO2 O2N NH R CH DNP-amino acid (Dinitrophenyl-amino acid) COOH + O Free Amino Acids Dansyl Chloride CH3 CH3 CH3 CH3 N N + O S H2N R O C C H O HCl Base O S Cl Dansyl Chloride 1-Dimethylaminonapthalene5-sulfonyl chloride H+ Dansyl amino acid + Free Amino Acids HN O R O C C H End Group Analysis (How Many Different Subunits?) C-Terminal Identification Reduction R NH CH _ COO LiBH4 R NH CH CH2OH H+ Amino Acids + Amino Alcohol Hydrazinolysis R1 NH CH O C R2 NH CH COO– H3N R O CH C NHNH2 hydrazides :NH2NH2 + R2 H3N CH COO– only unaffected aa Cleavage of Disulfide Bonds Oxidative Cleavage O HCOOH CH 2 S S CH 2 (Performic Acid) A Cystine B CH 2 SO3 – – O3S Cysteic Acid CH 2 Problem (Oxidation of Methionine to Methionine Sulfone) O CH2 CH2 S CH3 O Reduction and Alkylation 2 R–SH CH2 A S S "cystine" R–S–S–R CH2 CH2 A B HOCH2CH2SH -mercaptoethanol SH HS CH2 B Problem O2 CH2 SH HS CH2 A CH2 "oxidation" S S CH2 B A—B, A—A, B—B Solution HI R SH + ICH2CONH2 "iodoacetamide" RSCH2CONH2 Separation and Purification of Polypeptide Chains Sequence Polypeptide Chains • Specific peptide cleavage reactions • Separation and purification of peptide fragments • Sequence determination Hydrolysis Polypeptide Hydrolysis Amino Acids Acid Hydrolysis Seal N2 Protein 6N HCl e.g. AlaAspSer 110o 24 h 6N HCl 110°, 24h Individual Amino Acids Ala + Asp + Ser* Mechanism + H3N R1 CH R2 O C NH CH _ COO H+ H + R1 H3N CH O C + R2 NH CH H _ COO O C : : O H NH H+ O H H R1 H H3N CH COO– + R2 H3N CH COO– Problems • Complete destruction of Trp • Partial destruction of Ser, Thr, and Tyr • Deamination of Asn and Gln Deamination of Asn and Gln NH 2 O C + H3N O CH 2 H O C C N C C N C C R1 H O R2 Asn H+ (also hydrolysis of peptide bonds) Base Hydrolysis (Many Amino Acids Destroyed) (Racemization) B: H CH3 C NH C O L-amino acid CH 3 base H B CH 3 C NH H + :B C C O_ NH C O D-amino acid Enzymatic Hydrolysis Mild Conditions Many proteases and peptidases Specific and non-specific Problem: contribution of amino acids from hydrolysis of proteases Amino Acid Analysis (Automated) Ion-exchange chromatography High performance liquid chromatography Colorimetric Analysis Specific Peptide Cleavage Reactions Proteolytic Enzymes R1 . . . NH CH C O R2 NH CH C . . . O Cleave peptide bonds Specificity: R1 Specificity of Endopeptidases Table 5-3 Chemical Cleavage (Cyanogen Bromide) • • • NH CH C NH • • • • • • NH CH2 O C C CH2 O CH2 CH2 S CH CH3 N Br • • • NH CH CH2 • • • NH C + O + NH3 • • • CH2 peptidyl homoserine lactone peptide + S CH3 C N S Br O NH .. • • • H2 O + CH C NH • • • CH2 O CH2 C CH3 N methyl thiocyanate Separation and Purification of Peptide Fragments Sequence Determination: -Edman degradation -Mass Spectrometry Edman Degradation I R1 O N C S + H2N C C H Phenylisothiocyanate Base R1 O NH C NH C C NH S H PTC (Phenylthiocarbamyl–) Polypeptide Edman Degradation II R1 O NH C NH C C NH H S PTC (Phenylthiocarbamyl–) Polypeptide Anhydrous HF N R1 R2 + NH S H3N CH O Thiazolinone Derivative O C Polypeptide Edman Degradation III R1 N R2 + NH S H3N CH O Thiazolinone Derivative Mild Acid S C NH N C CH R O PTH (Phenylthiohydantoin) Amino Acid O C Polypeptide Electrospray Ionization Mass Spectrometry (ESI) Figure 5-16a part 1 Electrospray Ionization Mass Spectrometry (ESI) Figure 5-16a part 2 Electrospray Ionization Mass Spectrometry (ESI) Figure 5-16b Tandem Mass Spectrometry Figure 5-17 Organize Completed Structure • Ordering peptide fragments • Assignment of disulfide bond positions • Determine position of amides Ordering Peptide Fragments Generating Overlapping Fragments Figure 5-18 Ordering Peptide Fragments Overlapping Fragments Trypsin Tyr • Lys Glu • Met • Leu • Gly • Arg Ala • Gly • Lys CNBr Tyr • Lys • Glu • Met Leu • Gly • Arg • Ala • Gly • Lys Complete Amino Acid Sequence Tyr • Lys • Glu • Met • Leu • Gly • Arg • Ala • Gly • Lys Assignment of Disulfide Bond Positions Hydrolyze without breaking disulfides Reduce, alkylate, and identify linked fragments (disulfides) Assignment of Amide Positions Hydrolyze without breaking amides Hydrolyze fragments and measure NH3 (need fragments having a single Asn or Gln) Protein Evolution Evolution by Natural Selection Mutations Cytochrome c All look like this Table 5-5 part 1 Sequence Comparisons Provide Information on Protein Structure and Function • Homologous proteins: evolutionarily related proteins – Invariant residues – Conservative substitutions – Hypervariable positions • Neutral drift Phylogenetic Trees Depict Evolutionary History Figure 5-21 Proteins Evolve by the Duplication of Genes or Gene Segments Protein Families Can Arise through Gene Duplication • Orthologous proteins: homologous proteins with the same function in different species • Paralogous proteins: independently evolving proteins derived by duplication of a gene (globin family) • Pseudogenes Globin Family Figure 5-22 The Rate of Sequence Divergence Varies Figure 5-23