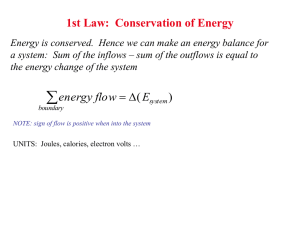
The Two-step Mechanism of Nucleation of Crystals from
advertisement
Kinetics and Thermodynamics of Multistep Nucleation and Self-assembly in Nanosize Materials Brussels, March 25-26, 2010 The Two-step Mechanism of Nucleation of Crystals from Solution Peter G. Vekilov, Oleg Galkin, Luis Filobelo, Weichun Pan, Anatoly Kolomeisky (Rice), Dimo Kashchiev (IPC), Vas Lubchenko Department of Chemical and Biomolecular Engineering, Department of Chemistry, University of Houston NIH, NASA, NSF, Welch Foundation The Goal: Crystals with “Just-right”… Number Polymorph Morphology Habit Size Size distribution Requires data on: Solution PChem Phase diagrams Metastable states Nucleation mechanisms Growth mechanisms Agglomeration … Crystallization and Nucleation Crystallization Nucleation Crystallization and Nucleation Crystallization Nucleation Concentration Crystallization as Sequential Transition along Two Order Parameters Structure Two-step mechanism: suggested by t W & F, T & O for critical point for L-L phase separation for proteins Everywhere else in phase diagram—classical crystal nucleation predicted P.R. ten Wolde, D. Frenkel,Science 277 (1997) 1975 V. Talanquer, D.W. Oxtoby, J. Chem. Phys. 109 (1998) 223 Solubility Temperature Classical viewpoint: direct nucleation along a “diagonal line” envisioned; … Gelation Binodal Spinodal Protein Concentration Concentration The Two-step Mechanism It may apply to all crystals (and other ordered solids) forming in solution Galkin, O. & Vekilov, P. G. (2000) Proc. Natl. Acad. Sci. USA 97, 6277 Vekilov, P. G. (2004) Crystal Growth and Design 4, 671 Solubility Temperature It operates in all areas of the phase diagram Structure … Gelation Binodal Spinodal Protein Concentration TL-L Clys = 50 mg/ml Clys = 80 mg/ml 0.4 Solubility Temperature Homogeneous Nucleation Rate J [cm-3 s-1] The Nucleation Rate 0.2 Gelation Binodal T L-L Spinodal Protein Concentration 0 5 10 15 20 Temperature T [°C] 25 30 Maximum in J(T) Exponential increase at intermediate DT’s; by weak decrease at higher DT’s T of maximum shifts with concentration Two steps: Which One is Rate Determining Concentration Structure … Critical level of ordering Critical size of ordered nucleus Rate of cluster formation J1 ~ J01exp(– DG1*/kBT) DG2* crystals Is J02 more important than DG2*? DG1* DGL-L DG2* dense liquid Is J01 more important than DG1*? solution * * Is DG1 > DG2 ? Free Energy G Rate of nucleation within clusters J2 ~ J02exp(–DG2* / kBT) DG1* DGL-L Nucleation Reaction Coordinate Nucleation of Dense Liquid Droplets T – TL-L = 0.7 oC 3.53 s 6.09 s 10.58 s 1.92 s 7.37 s T – TL-L = 1.3 oC 0.96 s • Number of droplets increases with time • Faster nucleation at higher DT’s Characteristics of nucleation regime of droplet generation Nucleation Rate of Dense Phase Droplets Droplets in Viewfield 400 M. Shah, et al., J. Chem. Phys. 121 (2004) 7505 300 200 • Number of droplets increases in time— nucleation regime Nucleation Rate = 4.3 x 109 cm-3s-1 • Nucleation rate ~109 cm-3s-1 significantly higher than rates of crystal nucleation ~ 0.1 – 1 cm-3s-1 100 0 0 2 4 6 Time [s] 8 10 Structuring of dense liquid quasi-droplet is the rate determining stage Equilibrium between solution and clusters: msolution = mclusters Dm(solution,crystal) = Dm(clusters,crystal) Other proteins: • Ferritin crystals grown at s = 4.2, where n* 1 • Are protein crystals always grown in spinodal regimes? -3 -1 • Spinodal – boundary between metastable and unstable two – phase areas Nucleation Rate J [cm s ] Why is the maximum in J(T) sharp? 10 0 10 -1 10 -2 • 10.0oC 12.6oC 15.0oC n* = 1 2.8 3.2 3.6 Supersaturation Dm/ kBT Spinodal can be defined from n* 1 L.F. Filobelo, et al., J. Chem. Phys. 123, 014904 (2005) Pre-exponential Factors and Barriers for Structuring 50 Liquidus or solubility of crystals Gelation line o Temperature [ C] 40 Solution-crystal spinodal 30 L-L coexistence 20 10 0 D.N. Petsev, et al., J. Phys. Chem. B 107 (2003) 3921 L.F. Filobelo, et al., J. Chem. Phys. 123 (2005) 014904 L-L spinodal -10 0 20 mm 100 200 Concentration [mg/ml] 300 Why is the maximum in J(T) sharp? 50 Liquidus Gelation line o Temperature [ C] 40 J(T) reaches sharp max at solution-crystal spinodal 30 L-L coexistence 20 10 0 Liquid-liquid (L-L) spinodal -10 0 100 200 Concentration [mg/ml] 300 Phenomenological Theory of Two-step Nucleation -1 -3 -1 Nucleation Rate J [cm s ] 012 0.5 u1 (T ) 1 1 u0 (T ) u0 (T )u 2 (T ) u 2 (T ) – mean first-passage time J = -1 , 2 – rate-limiting E U 2 exp( 2 ) k BT J U DG 1 1 exp( ) U0 k BT U 2 k2 C1 T (C1 , T ) 0.4 0.3 0.2 0.1 0.0 Viscosity inside dense liquid 0 1 C1 expk C1 exp E / k BT E 2 (T ) E* Te T 2 T T 2 1 e Te Tsp 2 Single adjustable k2 reproduces 3 complex kinetic curves W. Pan, et al., J. Chem. Phys. 122, 174905 (2005) 276 280 284 288 292 Temperature T [K] 296 300 T = 12.6 oC -3 -1 Nucleation Rate J [cm s ] Nucleation barrier on approach to spinodal 80 mg ml -1 50 mg ml 0.2 0.1 0.0 20 30 40 50 -1 Concentration C [mg ml ] 60 The Pre-exponential Factor in the Nucleation Rate Law DG * J J0 exp( ) kBT From experiments: J0 ~ 1010 cm-3s-1 From classical theory J0 ~ 1020 cm-3s-1 ??? R.P. Sear, J. Phys. Chem. B 110 (2006) 21944 E2 ) k BT J U DG 1 1 exp( ) U0 k BT U1 DG 1 exp( ) , cluster volume fraction U0 kBT U 2 exp( Low J0—due to nucleation within clusters Volume Fraction From phenomenological theory: 10 -7 0 50 100 150 200 Time of Monitoring [min] 250 The Two-step Mechanism for Other Crystals Glycine, urea B. Garetz, et al., Phys. Rev. Lett. 89, 175501 (2002) J.E. Aber, et al., Phys. Rev. Lett. 94, 145503 (2005) D.W. Oxtoby, Nature 420, 277 (2002) Charged colloid crystals M. E. Leunissen, et al., Nature 437, 235 (2005) NaClO3 R.Y. Qian, G.D. Botsaris, Chem. Eng. Sci. 59, 2841 (2004) NaCl nucleation from solution (MD simulation) D. Zahn, Phys. Rev. Lett. 92, 040801 (2004) Calcite nucleation L. Gower, Chem. Rev. 108, 4551 (2008) D. Gebauer, et al., Science 322, 1819 (2008) Theoretical Justification of Generality of 2step Mechanism J. Lutsko, G. Nicolis, Phys. Rev. Lett., 96 (2006) 046102 For protein molecules b c DG/kBT a c b a For small molecules e f DG/kBT d Reaction Coordinate Two-step barrier always lower than direct barrier f e d q(T) much stronger than R(T) contradicts 1-step nucleation and agrees with 2-step 0.6 1/R [s mm-1] Clusters and HbS Polymer Nucleation 0.4 0.2 0.0 D. Kashchiev, et al., J. Chem. Phys. 122 (2005) 244706 0 Polymers are perpendicular to plane of polarization of polarized light t=12 s n=1 t=15 s n=2 15 20 q [s] 90 25 60 30 150 180 0 330 210 240 300 800 270 Radius [nm] 600 O. Galkin, P.G. Vekilov, J. Mol. Biol. 336, 43 (2004) O. Galkin, et al., J. Mol. Biol. 365, 425 (2007) O. Galkin, et al., Biophys. J. 92, 902 (2007) P.G. Vekilov, Brit. J. Haematol. 139, 173 (2007) t=0 n=0 10 120 Dependencies of r, Vl and Nl of mesoscopic metastable clusters on C and T follow those of nucleated polymers Clusters are precursors for polymer nuclei 5 400 200 0 t=18 s n=3 o o 10 C o 25 C 15 C o 30 C 20 o 20 C 40 60 Time of Monitoring [min] t=21 s n=5 Aggregation Precedes Ordering in Biological Self-assembly Hemoglobin assembly—from 2 a-chains, 2 b-chains and 4 heme-moieties after translocation a- and b-chains associate prior to folding K. Adachi, et al., J Biol Chem 277, 13415 (2002) Hemes attach to a2b2 complex and then enter assigned slots G. Vasudevan, M. J. McDonald, Curr Protein Pept Sci 3, 461 (2002) Nucleation of prion-protein fibers—via a disordered toxic fluid-like cluster Molten Oligomer Nucleus Fibril R. Krishnan, S. L. Lindquist, Nature 435, 765 (2005) A. Lomakin, et al., Proc. Natl. Acad. Sci. USA 93, 1125 (1996) E. H.Koo, et al., Proc. Natl. Acad. Sci. USA 96, 9989 (1999) Density The Two-step Mechanism … What are the consequences for the nucleation kinetics? Structure What are the precursors above the L-L coexistence line? Does it offer new “handles” for control? Temperature Solubility Gelation Binodal Spinodal Protein Concentration 100 µm Crystals Do Not Nucleate Within Liquid Droplets t=4h t=6h t=8h t = 10 h What else may be precursor? t = 12 h t = 14 h t = 16 h Direct Observation of Clusters lumazine synthase 3 mg/ml 1.3 M phosphate 24 C Clusters shrink in height: Height [nm] Steps generated from clusters merge continuously with underlying lattice Clusters represent hidden dense liquid 200 200 100 100 0 0 0 2.5 5 7.5 10 Surface Coordinate [mm] 0 2.5 5 7.5 10 Surface Coordinate [mm] O. Gliko, et al., J. Amer. Chem. Soc. 127 (2005) 3433 W. Pan, et al., Biophys. J. 92 (2007) 267 O. Gliko, et al., J. Phys. Chem. B 111 (2007) 3106 Evidence for Mesoscopic Clusters in Protein Solutions Dynamic light scattering determinations oxy HbS C=169.6 mg/ ml deoxy HbS C=131.2 mg/ml 0.6 100 Lysozyme78 mg ml-1, 20 mM HEPES, pH = 7.8 10 0 Clusters 0.3 HbS molecules 0.0 1E-4 0.01 1 100 Delay Time [ms] Cluster size n100 nm Cluster lifetime >15 ms to 10 s 100 150 200 250 400 0 50 100 150 deoxy-HbS 1000 Steady volume 10-8—10-3 V: not crystals Fast decay rate indicates that clusters are hidden liquid 50 LuSy 8.1 mg/mL 1.3 M phosphate pH = 8.7 800 Radius [nm] g2() – 1 0.9 67.2 mg/ml 131.2 0.15 M phosphate pH = 7.2 100 0 50 100 150 Time of Monitoring [min] O. Gliko, et al., J. Amer. Chem. Soc. 127 (2005) 3433 The Angular Dependence Decay rate G2 [ms] 2.5 q = 4pn/l sin(q/2) 2.0 q– scattering angle Cluster peak 1.5 For freely diffusing clusters G2 = Dq2 1.0 0.5 0.0 0 200 400 q2 600 Loose network is anisotropic environment G2 ≠ Dq2 [mm-2] Freely-diffusing clusters G2 = Dq2 Their size – from Einstein-Stokes law D = kT / 6pR2 The Free Energy Cost of Higher Concentration 15 K /Rq ( /) /RT 0 H DG ~ 10 kBT -2 10 -4 -6 5 Fraction of protein in clusters: exp(-10) = 4 × 10-5 an overestimate Clusters must contain only a few molecules Cluster lifetime must be O(diff ) = 10 ms DG/NkBT L MwK/Rq DG dV D ( V ) -8 0 0 100 200 300 400 -10 -1 Protein Mass Concentration [mg ml ] Clusters must contain a chemical species with slow decay rate Open Question about the Clusters (a) -1 10 (b) -1 2 -3 10 -5 10 80 mg ml o 57 o 77 o 96 o 116 o 140 150 mg ml -7 10 (c) -1 2 -3 10 (d) 334 mg -1 228 mg ml -5 -7 -3 10 -1 10 1 10 sin2(q/2) [ms] 3 10 -5 10 -3 10 Cluster volume fraction less than suggested by equilibrium free energy Clusters are fluid internally Complicated dependence of cluster volume fraction on protein concentration ml-1 10 10 -5 10 Macroscopic lifetimes Cluster size independent of protein concentration -1 10 Mesoscopic size -1 10 1 10 sin2(q/2) [ms] 3 10 Cluster shoulder--nonexponential at high concentrations Microscopic Scenario and the Cluster Radius Clusters consist of offequilibrium mixture of monomers and oligomers, kinetically stabilized Cluster radius R is determined by the decay rate and diffusivity of the oligomers: R (D2/k2)1/2 Since R ≃ 100 nm, oligomer lifetime k2-1 10 ms Cluster radius does not depend on concentration The Cluster Lifetime -1 2 10 -3 10 -1 228 mg ml -5 10 -7 10 -5 10 -3 -1 10 10 1 10 3 10 -1 2 10 R2 2 D1 -3 10 Cluster size fluctuates with characteristic time The imprint of these fluctuations on the correlation functions scales with q2 334 mg ml-1 -5 10 ≈ 10 ms is the lower bond of cluster lifetime -7 10 -5 10 -3 10 -1 1 10 10 2 *sin (q2) [ms] 3 10 The Oligomer Mechanism: Hydration 80 40 -3 10 2() Free Energy [kJ/mol] -1 10 0 -5 -40 10 2 4 6 8 10 12 14 Separation [Å] -7 10 -1 150 mg ml 20 mM Hepes, pH 7.8 0.1M acetate, pH 4.5 20 mM phosphate, pH 7.8 -5 10 -3 10 -1 1 10 10 2 *sin (q2)[ms] Water structuring leads to secondary minima in interaction between nanoscopic solutes Small ions, i.g., HPO42- , are known to disturb the hydration shell 3 10 The Oligomer Mechanism: Domain Swapping -1 80 mg ml No urea 0.2 M urea 0.5 M 1M -1 10 -3 2( ) 10 -1 150 mg ml No urea 0.2 M urea 0.5 M 1M -5 10 -7 -3 10 -1 1 10 10 2 *sin (q/2) [ms] 3 10 Attraction between solvent exposed hydrophobic residues, “domain swapping” Urea can be used to control degree of unfolding -5 -3 10 10 -1 10 1 10 3 10 *sin2(q/2) [ms] 0.5 M urea 0.5 M urea no urea 10 0 -2 -4 -6 -8 5 DG/NkBT -5 10 MwK/Rq 10 -10 0 100 200 300 400 500 -1 Lysozyme Concentration [mg ml ] Summary and Conclusions Assembly of ordered arrays crystals, oligomers, fibers, etc. is preceded by association into disordered clusters The precursor is a metastable mesoscopic liquid cluster Rate of crystal nucleation is determined by structuring of dense quasidroplet Polymorph selection is determined by kinetics factors rather than by high barriers The low volume fraction of the nucleation precursors delays nucleation by ~ 1010 Understanding and control of nucleation in solution requires insights into the solution physicochemical mechanism nano- and mesostuctures So What? Clusters are needed for nucleation of crystals. To enhance clusters: moderate intermolecular attraction or repulsion proper water structure around the protein molecules Crystal nucleation occurs in a spinodal regime g is not important Simpler picture of nucleation and role of additives Heterogeneous particles may affect polymorph selection via structural similarity