Nuclear gene expression 1
advertisement

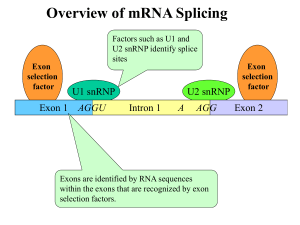
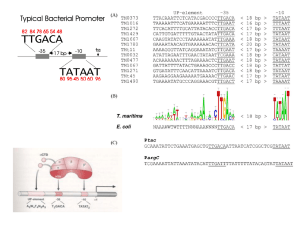
Plant Nuclear Gene Expression & Regulation A lot of steps to regulate: 1. Transcription* 2. Capping 3. 3' maturation, cleavage & polyadenylation 4. Splicing* 5. Transport to Cytoplasm 6. Stabilization/Destabilization of mRNA* 7. Translation* * have the most regulation. Likely order of events in producing a mature mRNA from a pre-mRNA. Transcription: 3 DNA-Dependent RNA Polymerases 1. Pol I - synthesizes 45S rRNA precursor, found in nucleoli (45S18S, 28S, 5.8S rRNAs) [S refers to rate of sedimentation (Fig. 6.33 in Buchanan), approx. equivalent to size of macromolecule] 2. Pol II - synthesizes mRNA precursors, some snRNAs 3. Pol III- synthesizes 5S rRNAs, tRNAs, small nuclear RNAs (snRNAs) All 3 polymerases are multi-subunit; have some large, unique subunits; and 5 small, shared subunits (at least in yeast). Relative cellular RNA abundance • Ribosomal RNAs (rRNAs) • Transfer RNAs (tRNAs) • Messenger RNAs (mRNAs) The rest (~3%): • Signal recognition particle (SRP) RNA • Small nuclear RNAs (snRNAs) • Small nucleolar RNAs (snoRNAs) • Micro RNAs (miRNAs) ~ 90% ~ 5% ~ 2% RNA Polymerase II 1. 2 large subunits have regions of homology with ß and ß’ subunits of E. coli RNAP. 2. Largest subunit is phosphorylated on its COOH-terminal domain (CTD) – Phosphor. needed for transition from initiation elongation – CTD also interacts with other proteins 3. Does not bind DNA by itself, requires other proteins to bind promoter first! TFII – transcription factors for RNA Pol II RNAPII – RNA Pol II Fig. 6.30, Buchanan et al. RNAP II Promoters • Class-II promoters have 4 components: 1. Upstream element(s) 2. TATA Box (at approx. –25) 3. Initiation region (includes the first transcribed nt, +1) 4. Downstream element 1. 2. 3. 4. Many class II promoters lack 3 and 4; a few lack 2. TATA Box of Class II Promoters • TATA box = TATAAAA • Defines where transcription starts • Also required for efficient transcription for some promoters • Bound by TBP – TATA box binding protein (in complexes like TFIID) Upstream elements: Class II promoters Found in many class II promoters: 1. GC boxes (GGGCGG and CCGCCCC) – Stimulate transcription in either orientation – May be multiple copies – Must be close to TATA box 2. CCAAT box – Stimulates transcription – Binds CTF (Cat-box transcription factor) Enhancers and Silencers 1. Enhancers stimulate transcription, while Silencers inhibit. 2. Orientation-independent – Flip 180 degrees, still work 3. Position-independent (mostly) – Can work at a distance from promoter core – Enhancers have been found all over 4. Bind regulatory transcription factors Transcription factors for Class II promoters 1. Basal factors: required for initiation at most promoters; interact with TATA box. 2. Upstream factors: bind common (consensus) elements upstream of TATA, including proximalpromoter elements (e.g., CCAAT box); increase efficiency of initiation. 3. Inducible (regulated) factors: work like upstream factors but are regulatory (produced or active only at specific times/tissues); interact with enhancers or silencers. Assembly of the RNA Pol II Initiation Complex = basal factors + RNAP II TFIIF delivers Pol II TFIIH PO4ylates the LS of Pol II, allowing it to escape the promoter. Fig. 7.45, Buchanan et al. Eukaryotic Transcription Factors: Structure • Mostly about factors that bind USEs: 1. Modular structure: – DNA-binding domain – Transcription-activating domain 2. Can have > 1 of each type of module 3. Many factors also have a dimerization domain (some can form heterodimers). DNA-binding domains 1. 2. 3. 4. Zinc – containing modules Homeodomains (conserved amino acid seq.) bZIP and bHLH motifs AP2 (mainly in plants) (not an exhaustive list, just what might be on the test!) Activation from a Distance: Enhancers • 3 possible models Factor binding induces: 1. Supercoiling of the promoter DNA 2. Sliding of the complex to the promoter 3. Looping out of DNA between enhancer and promoter 3 Models of possible enhancer action. Chromatin Modification • Transcription can also be regulated by modifying chromatin (histones); highly transcribed genes have less condensed chromatin. • Basic unit of chromatin is the nucleosome: 1. 4 different histones in the core (H2a, H2b, H3, H4 x 2 = octamer) 2. 146 bp of DNA wrapped around core 3. Histone H1 on outside H2 A H3 H2B H1 H4 H2 B DNA Nucleosome core = octamer of histones (2 each of H2A, H2B, H3, H4) + 2 wraps (145 bp) of DNA Packing ratio ~5 Histones can be modified (for chromatin remodeling) Histone acetylation (right) causes localized unpacking of nucleosomes, which enhances factor binding to DNA. De-acetylated histones (left) bind DNA more strongly, and the nucleosomes condense into a solenoid; this inhibits factor binding to DNA targets. Fig. 7.49 Buchanan et al. In Vivo Studies • Promoters of active genes are often deficient in nucleosomes SV40 virus minichromosomes with a nucleosomefree zone at its twin promoters. Can also be shown for cellular genes by DNase I digestion of chromatin – promoter regions are hypersensitive to DNase I. Fig. 13.25 Post-Transcriptional Processes 1. Capping 2. 3’ end formation (not much regulation of the above steps) 3. Splicing – alternative splicing 4. Translation – regulate initiation step Cap Functions • Capping also includes methylation of the ribose (2-OH) on nt #1 and sometimes #2. • 1. 2. 3. 4. Cap functions: Protection from 5 exoribonucleases Enhances translation in the cytoplasm Enhances transport from the nucleus Enhances splicing of the first intron (for some pre-mRNAs) 3’ end Processing & Polyadenylation Mechanism • Transcription extends beyond mRNA end • Transcript is cut at 3’ end of what will become the mRNA • PolyA Polymerase adds ~250 As to 3’ end • “Extra” RNA degraded 3' End Formation CIS (elements) • AAUAA is the key signal in higher plants, its found ~20 nt from the polyA-tail. – Other sequences 5' to the AAUAA also important. TRANS (factors) • 3' end formation requires at least: – an endonuclease & recognition factors – a poly(A) polymerase (PAP) – a poly A-binding protein (PAB)