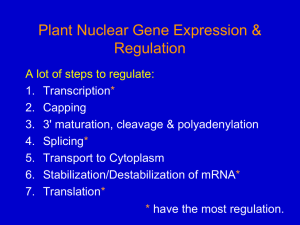
Gene Expression in Eukaryotes
advertisement

FUNDAMENTALS: 11:00-12:00 DATE: Monday, SEPTEMBER 13, 2010 PROFESSOR: RYAN I. II. III. IV. V. VI. VII. GENE EXPRESSION Scribe: BO BRADFORD Proof: ROSS ISBELL Page 1 of 7 EUKARYOTIC RNA POLYMERASE [S2] a. Eukaryotes have three dependent RNA polymerases I, II, III b. All are large multi-unit proteins c. 2 larger sequences are similar to the Beta & Beta prime of the E. coli RNA polymerase d. The catalytic site is conserved e. All 3 polymerases interact with general transcription factors-GTF- They use these to recognize their promoters f. Use alpha-amanitan in order to see what genes where transcribed by RNA POL ii. g. Most mRNA’s of proteins are transcribed via RNA Pol II RNA POLYMERASE II INHIBITOR- ALPHA AMANITAN [S3] a. Bicyclic octopeptide, two cyclic rings with 8 peptides b. POL II is the most sensitive to this enzyme followed by POL III, then POL I i. POL II > POL III > POL I YEAST RNA POLYMERASE II SUBUNITS [S4] a. Chart that shows the subunits of the yeast RNA POLYMERASE II b. Pol subunits are called i. R=RNA ii. P= Polymerase iii. A= subunit 1; B=subunit 2; C=subunit 3 c. These are all arranged accordingly by their overall size d. The largest subunit- RPB1, shares homology with the E. coli beta prime subunit. It has a region called the carboxy terminal domain which is a repeating heptapeptide e. RPB2- has the nucleotide triphosphate binding site & has homology with the beta subunit of E. coli f. RPB3 has homology with the alpha subunit & RPB4 with the sigma subunit g. Some subunits are shared between all three polymerases, and some are specific only to RPB2 h. Each of the specific Pol I, II, III have unique subunits 14, while some others are shared TRANSCRIPTION FACTORS [S5] a. All three RNA POL I, II, III interact with their promoters via a protein: protein interactions & protein: DNA interactions b. These proteins are transcription factors that recognize & initiate transcription @ specific promoter sequences. c. Some transcription factors-for example TFIIIA & TFIIIC for RNA POL III bind specifically to the recognition sequences WITHIN the coding regions. THEY DON’T ALWAYS BIND UPSTREAM. HELIX-TURN-HELIX MOTIF [S6] a. Type of transcription factor b. Found very commonly in Prokaryotes, and sometimes eukaryotes c. Examples in Prokaryotes= TRYP REPRESSOR, LAC REPRESSOR, CATABOLITE ACTIVATOR PROTEINS d. Examples in Eukaryotes= ANTP (atinopedia) e. All have two alpha helices connected by a turn f. These bind DNA’s by placing one of the alpha helices within the major groove of the DNA & the second helix holds the motif in place. g. Usually bind as DIMERS, allowing for dyad symmetry at the binding site. ZINC FINGER MOTIF: C2H2 CLASS [S7] a. C=cysteine H= Histidine b. Another type of transcription factor c. Have 4 amino acids coordinating a zinc atom d. Amino acids that extend between the cysteine & histidine residues form the actual “finger” e. This is the secondary structure: Half of the molecule forms an alpha helix, and the finger contacts the DNA in the major groove f. Most fingers will recognize 5 base pairs within the loop, there are 7/8 important side chains on the amino acids that will contact the 5 bases on DNA. Most zinc finger proteins have multiple fingers g. Each finger will add 3 bases to the recognition sequence h. Can be manufactured to bind to almost any type of DNA, can be used to change genes, as well as mutate them. i. Thousands of these within our genome, NOT ALL bind to DNA, but a lot do. BASIC REGION- LEUCINE ZIPPER MOTIF [S8] FUNDAMENTALS: 11:00-12:00 DATE: Monday, SEPTEMBER 13, 2010 PROFESSOR: RYAN a. b. c. d. e. GENE EXPRESSION Scribe: BO BRADFORD Proof: ROSS ISBELL Page 2 of 7 A.k.a= b-zip Basic domains that contact the DNA within the major groove. Alpha helical region called the leucine zipper that has a leucine present at ever 7 amino acids So you have an aliphatic helix- one side is polar & the other is non-polar (the leucine rich region) These differences in polarity allow for two b-zip’s to join with one another via DIMERIZATION; the two nonpolar surfaces zip right up with one another in order to isolate themselves. f. The dna binding region is at the top (it would be my hands) where as the zipper would be at the bottom ( my forearms). g. Another type of transcription factors VIII. Bzip TRANSCRIPTION FACTOR [S9] a. Schematic showing the binding of a b-zip bound to DNA double helix b. Finger on top of the helix and another going behind within the major groove c. You can have homodimers of 2 leucine zippers or heterodimers of leucine zippers i. 2 different b-zips that are adjoined by the same leucine zipper but recognize different DNA regions IX. GENERAL TRANSCRIPTION FACTORS-GTF [S10] a. GTF’S help to position RNA Polymerases at the transcription initiation sites, forming the preinitiation complex- PIC ( this is the polymerase + all factors) b. Many GTF’s that are associated with RNA Polymerase II initiate transcription from the TATA boxhave been identified. a. TFIIA, TFIIB, TFIID, TFIIE, TFIIF, TFIIH ( TF=transcription factor, II= RNA polymerase II & a/b/d…= subunit type). b. Each one of these complexes have been purified in identified in RNA Pol II c. ALL OF THESE TRANSCRIPTION FACTORS ARE NEEDED IN ORDER TO FORM A PREINITIATION COMPLEX- PIC @ POLERMASE II PROMOTER REGION X. GENERAL TRANSCRIPTION FACTORS- GTF [S11] a. TFIID subunit of the RNA Pol II is the largest & consists of a TATA-box binding protein (TBP) there are 8-10 associated factors (TAFII) that function with TFIID c. TFIID= TBP & A BUNCH OF TAF’S d. TBP= universal transcription factor, it associates with the promoters FOR ALL THREE POLYERMASES- I, II, & III; even for promoters WITHOUT A TATA BOX (it will even bind to promoters without a TATA box). e. Binding of TAFII’S extends the interactions of TFIID f. TFIID has two primary roles a. FOUNDATION for the transcriptional PIC complex b. PREVENTS nucleosomes from encroaching on the area & making the promoter disappear. If this is a gene you want to continuously transcribe- you don’t want the promoter to vanish- you want it to stay exposed & readily available. ACTS AS AN ANTAGONIST TO HI (subunit of histone protein). XI. TBP IS USED BY ALL 3 RNA POLYMERASES [S12] a. TBP is a subunit important GTF for all 3 polymerases b. It’s called TFIID for Polymerase II c. Called SL1 for Pol I d. TFIIIB for Pol III e. DOESN’T always bind to the TATA Box f. The GTF’s that contain TBP act as positioning factors for their respective polymerases at the transcription start site XII. YEAST TATA BOX BINDING PROTEIN [S13] a. Structure of the yeast tata binding protein b. As opposed to ALL other transcription factors we’ve talked about, tata binding protein binds in the MINOR GROOVE of DNA c. When it binds it bends the DNA by a 120 degrees, so it shifts the duplex d. Has been described as a saddle for a polymerase to jump on and start riding (Hardy..Har..Har..) XIII. PROMOTERS FOR RNAP I [S14] a. RNA Pol I transcribes genes for the large rRNA precursor- the 18s, 5.8s, 28s ribosomal RNA’s are all made from one large ribosomal precursor FUNDAMENTALS: 11:00-12:00 Scribe: BO BRADFORD DATE: Monday, SEPTEMBER 13, 2010 Proof: ROSS ISBELL PROFESSOR: RYAN GENE EXPRESSION Page 3 of 7 b. Around 200 hundred copies of these genes within our genomes; one of the most highly transcribed genes c. DOESN’T encode for proteins but rather ribosomal RNA’s d. Eukaryotic cells have a specific RNA Pol I to transcribe JUST THESE GENES e. RNA Pol I promoters have two components a. CORE ELEMENT= -45+20 ( this is where the SLI w/ tbp binds) b. UPSTREAM ELEMENT= -156 -107 ( this is where an upstream binding factor- UBF binds) XIV. TRANSCRIPTION OF rRNA GENES BY RNA POLYMERASE [S15] a. Pictured are two RNA genes b. Red=non-transcribed spacers that occur one after another within the genome c. RNA polymerase binds & transcribes between these red regions making a large 45s pre-ribosomal RNA that will eventually be processed into three components- 5.8s, 18s, 28s*** d. All three smaller subunits- 5.8s,18s, 28s are incorporated with other proteins to form the 40S &60S subunit; 5.8S + 28S + 50 proteins= 60S 18S + 30 proteins= 40S e. RNA Pol I transcribes ONLY ribosomal RNA genes XV. PROMOTERS FOR RNAP III [S16] a. Transcribes various SMALL rRNA’s- 5S rRNA, tRNA, U6 snRNA (small nuclear RNA’s-involved with splicing) b. 5S rRNA & tRNA genes have Class III promoters c. Small nuclear RNA’s- U6 snRNA- have promoters that resemble RNA Pol II promoters-however if you exposed these genes to alpha-amanitan, the genes are still transcribed readily- because they are transcribed by Pol III, not II; even though U6 snRNA appears similar to RNAP II promoters & have TATA boxes. d. 5S Rna promoter is within the coding region of the gene, instead of being upstream like the others we’ve discussed. e. tRNA’s promoters contain two elements XVI. PIC ASSEMBLY FOR RNA POL III GENES [S17] a. box a & box b are within the coding region-the binding sites for tRNA b. we have transcription factors TFIIIC, TFIIIB, followed by Pol III c. TFIIIC binds first, bringing in TFIIIB that has the TATA Binding protein-TBP-as one of its subunit, and finally Pol III is brought in forming the PIC d. the schematic to the right is similar, but it is the 5S RNA gene a. here we have TFIIIA followed by TFIIIC, TFIIIB with TBP, and finally Pol III e. both of these are class three promoters, they are located entirely within the coding region XVII. RNA POLYMERASE II GENERAL TRANSCRIPTION FACTORS [S18] a. TFIID is composed of TBP & TAF’s. The tbp recognizes to the core promoter at the TATA site but can also bind without TATA boxes. TBP recruits TFIIB b. TAF’s, part of TFIID also recognize a core promoter. It can do this without the TATA Box. A lot of TAF’s are involved with Positive/ Negative Repression c. TFIIA- has 3 subunits stabilizes TFIID @ promoter binding d. TFIIB- recruits RNA Pol II which is also associated TFIIF, which assists the binding with the promoter e. TFIIF- associated with Pol II & assists binding with the promoter f. RNA Pol II- has 12 subunits & is responsible for enzymatic synthesis of RNA, and the recruitment of TFIIE g. TFIIE- has the ATPase activity, this is where energy comes from for transcription. Also recruits TFIIH h. TFIIH- is a helicase that melts away the promoter region to open up the DNA. It is also a kinase that aids in promoter clearance by phosphorylating the carboxy terminal domain within the large subunit of the RNA Pol XVIII. TBP- ASSOCIATED FACTORS (TAF’S/ TAFII’S) [S19] a. TAF’s are important for helping TBP to bind to promoters that lack TATA boxes b. Different TAF’s in different cells! c. Hypothesized that this high Molecular Weight complex can assemble in one single step d. The TAF’s he described were result of in vivo reactions XIV. EUKARYOTIC RNA POL II TRANSCRIPTION [S20] a. these diagrams explain how these transcription factors play a role in the formation of the PIC b. TFIID-that has TBP & TAF’s- will bind to a TATA box on the promoter c. TFIIA stabilizes this structure while TFIIB comes in & helps orient TFIID with initiation site FUNDAMENTALS: 11:00-12:00 Scribe: BO BRADFORD DATE: Monday, SEPTEMBER 13, 2010 Proof: ROSS ISBELL PROFESSOR: RYAN GENE EXPRESSION Page 4 of 7 XV. EUKARYOTIC RNA POL II TRANSCRIPTION [S21] a. Once TFIIB is bound, TFIIF is bound to the RNA Pol. and helps guide the Pol to the TFIID/A/B on the promoter region XVI. EUKARYOTIC RNA POL II TRANSCRIPTION [S22] a. two large subunits of RNAP II interact with promoter DNA;\: CTD tail (unphosphorylated form) of RNAP II is in direct contact with TFIID b. TFIIE is finally brought in & then TFIIH phosphorylates the carboxy terminal domain & the polymerase begins elongating c. We need this whole PIC to be situated correctly to start transcription & the TFIIH phosphorylates the carboxy terminal domain & actually BEGINS transcription. After transcription begins, much of the PIC is left behind as elongation progresses XVII. EUKARYOTIC RNA POL II TRANSCRIPTION [S23] a. Again, TFIIH phosphorylates the carboxy domain & elongation process can begin b. PIC positioned correctly/accurately is critical XVIII. CARBOXYL TERMINAL DOMAIN (CTD TAIL) [S24] a. this is what gets phosphorylated for transcription elongation b. there are a lot of tyrosine, serines, that can be phosphorylated by TFIIH & it’s kinase function. c. The mouse has 52 repeats of this heptapeptide sequence listed on the slide XIX. RNA POL II PROMOTERS [S25] a. TATA box is where the TFIID will bind with the Tata binding protein b. Initiator box is highlighted-this is where the +1 start of transcription site is c. You can have upstream & downstream elements that also have transcription factor binding sites, that can bring down other factors to influence transcription XX. EUKARYOTIC TATA BOX [S26] a. start site of transcription usually always begins with a purine at the +1 start site b. in this diagram we have beta globin rings c. the initiator nucleotide binding site prefers a purine at initiation site- JUST AS IN PROKARYOTES d. TATA are the most highly conserved e. NOT ALL GENES HAVE TATA BOXES (a lot of Pol II’s, don’t)- they rely on other proximal/distal elements XXI. EUKARYOTIC PROMOTER REGIONS [S27] a. eukaryotic gene promoters are highlighted here in the schematic b. transcription factor binding sites can be repeated multiple times c. the top bar is the DNA SV40 early region promoter where you have a lot of these G/C rich binding sites that will bind the transcription binding factor SP1. d. Thymidine Kinase- the lower bar- has a bunch of different binding sites for TATA binding protein, SP1, CAAT box, or Octamer protein XXII. RNA POLYMERASE II PROMOTER CONSENSUS SEQUENCES [S28] a. chart that represents a series of the DNA binding sequences b. TATA box has a sequence of TATAAAA, the factor that binds it is the TATA binding protein & it’s size is listed as well as it’s abundance c. CAAT Box, G/C box also listed XXIII. PROXIMAL CONTROL OF ELEMENTS OF GENES [S29] a. beta globin gene schematic b. there are three exons, two introns, and transcription starting site shown above at the top of the slide & poly-a- addition site where transcription is eventually terminated c. also have promoter elements upstream & downstream d. within each individual element there are multiple components ;The beta globin promoter has a TATA box, a GATA box, an EKLF binding site- called a CAC box- another GATA box…etc. e. all of these genes have multiple binding sites for different transcription factors to bind f. The sum total of EVERYTHING THAT BINDS TO THE SEQUENCES results in transcription g. If this cell lacked THE GATA box, transcription can no longer be activated h. These are additional transcription factors that bind IN ADDITION to the other factors we’ve already discussed…they bind & keep these sites exposed & capable of being transcribed. XXIV. COORDINATE REGULATION VAI RESPONSE ELEMENTS [S30] a. this explains how we coordinate the regulation of multiple genes at the same time b. in prokaryotes we accomplished this by utilizing different sigma factors at different times, or catabolite repression (when glucose was low) FUNDAMENTALS: 11:00-12:00 Scribe: BO BRADFORD DATE: Monday, SEPTEMBER 13, 2010 Proof: ROSS ISBELL PROFESSOR: RYAN GENE EXPRESSION Page 5 of 7 c. In eukaryotes we can coordinate a regulation via RESPONSE ELEMENTS. These are DNA sequences that can bind specific transcription factors & these factors are activated in response to a specific environmental event- hormones, metallic substances, etc. d. Response elements are simply a dna sequence within the promoter of a given gene that will bind to a transcription factor & help bring in the Polymerase e. They are transacting factors XXV. RESPONSE ELEMENTS: COORDINATE REGULATION [S31] a. this is the metallothionein gene b. found in intestinal cells & becomes up regulated when exposed to heavy metals… c. called MRE’s=Metal Response Elements d. when exposed to cadmium for example, this will induce the expression of the metallothionein gene by increasing the transcription factor that binds to the MRE. e. Similarly, the Metallothionein gene can also be induced by stress; cortisol levels increase as a result, cortisol binds the glucocorticoid response element (GRE) thus activating transcription of Metallotheionein gene f. BLE=basal elements, are responsible for basic, low levels of expression-with the methallothionein factors you’ll increase expression XXVI. COORDINATE REGULATION BY HORMONES/ STEROID RECEPTORS [S32] a. shows a series of hormone receptors- ESTROGEN, PROGESTERONE, GLUCOCORTICOID, THYROID, RETINOIC ACID RECEPTOR b. all have similar structure c. all have DNA binding domain shown in orange in the middle; in the purple box is a hormone binding domain- each one binds to it’s respective hormone d. also have variable region that can serve as the activation domain. XXVII. STEROID RECEPTORS [S33] a. cartoon of what happens with the glucocorticoid receptor b. normally, receptor is located within the cytoplasm & inhibited by another complex-the inhibitory protein complex c. stress increases cortisol levels; cortisol binds to hormone binding sites causing a change in the structure of the protein causing the release of the inhibitory complex d. now activated glucocorticoid receptor with bound hormone now travels from the cytoplasm to the nucleus & bind to GRE elements XXVIII. ACTIVATION OF TRANSACTING FACTORS [S34] a. many ways to activate different transcription factors b. glucocorticoid example in slide 33 combines some of these mechanisms together c. you can increase transcription factor levels by: a. increasing protein synthesis/expression b. bind a ligand-glucocorticoid example c. phosphorylate an inactive protein d. add an additional subunit that activates it e. unmask an inhibitory protein-glucocorticoid example XXIX. CONTROL OF CELLULAR DIFFERENTATION BY TF’S [S35] a. can control differentiation by controlling different transcription factors b. embryonic cell undergoes a cell division and cell on the right begins expressing transcription factor I c. cells on the right in next division begin to express II & III d. finally, after a few cell divisions you have 8 cells with a variety of 5 different transcription factors within the cells e. each cell can express a unique set of genes based upon where the factors are binding XXX. ENHANCERS [S36] a. composed of multiple transcription factor binding sites b. enhance the level of transcription, usually act at a distance c. are orientation independent d. if they bind factors that repress transcription, then an enhancer is called a SILENCER XXXI. ENHANCERS [S37] a. all covered in previous slide XXXII. ENHANCERS: ACTION AT A DISTANCE [S38] a. schematic of an enhancer FUNDAMENTALS: 11:00-12:00 Scribe: BO BRADFORD DATE: Monday, SEPTEMBER 13, 2010 Proof: ROSS ISBELL PROFESSOR: RYAN GENE EXPRESSION Page 6 of 7 b. enhancer region binds to the activator protein (only one shown here-can be multiple), this assists in pulling in the RNA Polymerase and all associated general transcription factors & help it recognize the transcription initiation site-form the pre- initiation complex c. in the absence of the enhancer, this (RNA Pol) has a harder time binding to the promoter & transcription is decreased d. enhancer is critical to gene expression XXXIII. INSULATORS/ BOUNDARY ELEMENTS [S39] a. same locus control region that contains the enhancer also has these two functions b. works as an INSULATOR & BOUNDARY ELEMENT c. INSULATOR= blocks gene A from being enhanced by the same enhancer d. BOUNDARY ELEMENT- blocks the encroachment of heterochromatin form neighboring loci; heterochromatin is highly dense chromatin that isn’t expressed. e. Insulator boundary elements will be binding factors that keep everything between these two insulator elements open & accessible to transcription factors. In Erythroid ells this beta gene is expressed, in all other cells this beta gene isn’t expressed XXXIV. HYPOMETHYLATION VS. HYPERMETHYLATION [S40] a. a more permanent way to SILENCE GENE expression is DNA methylation b. schematic shows a DNA sequence upstream of the gene & when it’s being transcribes it’s is relatively HYPOMETHYLATED c. if factors are no longer produced & readily available, then other factors can come in and methylate the DNA-once methylated, another protein that recognizes methyl-cytosine can come in and bind bringing another complex that will remodel the region & close it off to transcription. d. This is a way to TURN OFF TRANSCRIPTION OF CERTAIN GENES. XXXV. SILENCING: HISTONE DEACETYLATION (HDAC) [S41] a. HDAC- wraps DNA up into a more inaccessible form of chromatin b. These deacetylate amino groups on the histone tails making them more positive. c. When histone tails are not acetylated the tails are more positively charged & this allows interaction with the DNA & neighboring histones-further condensing things d. HDAC result in more heavily condensed chromatin XXXVI. HISTON ACETYLATION AND DEACETYLATION [S42] a. does the opposite of HDAC b. HAT= HISTONE ACETYL TRANSFERASES c. Take positively charged side chains and adds an acetyl group- the HDAC’s remove these acetyl groups & leave a positive charge. This positive charge-CONDENSES THINGS FURTHER. d. When you acetylate the side chains, you open things up. XXXVII. HISTONE CODE [S43] a. these reactions- acetylation, methylation, phosphorylation, and ubiquitination occur at multiple places on the arms of histones b. nucleosomes with the DNA wrapped around them have a ton of lysines that can be potentially acetylated & opening them up. HDAC’S can make these more condensed c. Histone code= sum total of all reactions that take place on the histones, by looking at the sum total of the actions upon these histones, we can determine if a gene is in fact expressed or not. XXXVIII. CHROMATIN REMODELING [S44] a. require atp energy & can take histone octet and open it up making it accessible to transcription factors. b. Remodeling complex A-helps add factors where as Remodeling complex B- closes the octet back up again. XXXIX. GENE ACTIVATION BY CHROMATIN REMODELING [S45, S46, S47] a. the next few slides explain how we convert heterochromatin into euchromatin b. solenoid is condensed further with other proteins bound, transcription factor binds to the solenoid via it’s binding element c. Next, a HAT is recruited by the transcription factor and some histone tails become acetylatedfurther opening this region d. Afterwards, a DNA nucleosome remodeling complex along with ATP open the complex even further e. More transcription factors come in and bind f. This continues until the complex is open adequately, then the general transcription factors are recruited to bind g. FINALLY- RNA polymerase is recruited and now transcription of the gene can occur FUNDAMENTALS: 11:00-12:00 Scribe: BO BRADFORD DATE: Monday, SEPTEMBER 13, 2010 Proof: ROSS ISBELL PROFESSOR: RYAN GENE EXPRESSION Page 7 of 7 XXXX. GENETIC CLONING a. this is the experiment of the cloning of a cow, the cow is the egg donor b. take an unfertilized egg from the cow, extract the haploid genetic material with a needle, now you have an egg without DNA- it is now just a bag of chromatins that can remodel c. take a donor cell-any somatic cell- epithelial cell from oviduct d. uses the epithelial cell & fuses it with the given cell to form a diploid nucleus & insert it into the empty bag of protein that can remodel things. e. Once you do this you have a reconstructed zygote & the epithelial nucleus that was making all factors it needed in order to be an epithelial cell but now in a matter of hours, you can turn this cell into a reconstructed zygote & implant it into a female cow to birth a calf f. In a matter of hours the transcription factors, chromatin remodeling complexes, all histone modifying enzymes located in the egg are able to transform the epithelial cell into a zygote 53:03