Document
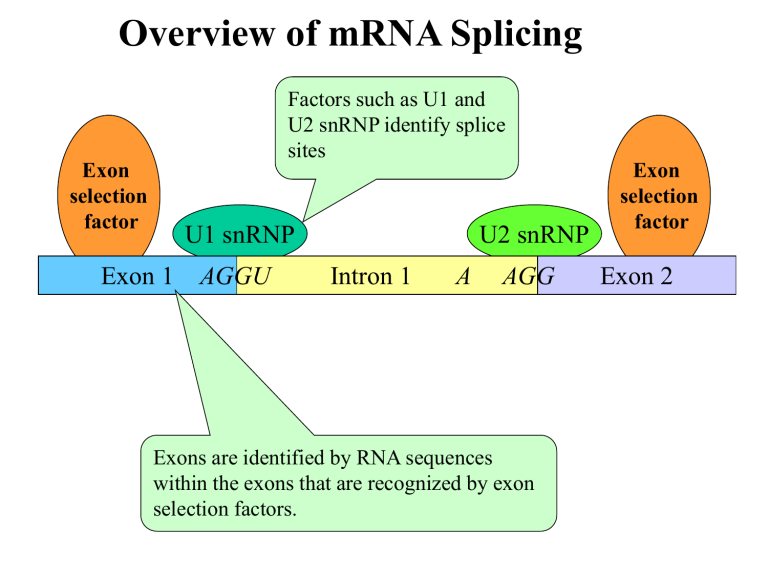
Overview of mRNA Splicing
Exon selection factor
U1 snRNP
Exon 1 AGGU
Factors such as U1 and
U2 snRNP identify splice sites
Intron 1
U2 snRNP
A AGG
Exon selection factor
Exon 2
Exons are identified by RNA sequences within the exons that are recognized by exon selection factors.
Beta globin splice mutations are one cause of beta thalassemia
EXON1 INTRON1 PHENOTYPE
AG GT AGT CONSENSUS
G
GCCAG GTTGGTAT NORMAL
GCCAG A TTGGTAT
0 (no beta chains)
GCCAG T TTGGTAT
0 (no beta chains)
GCCAG GTTG
GCCAG GTTG
T
C
TAT
TAT
+ (some beta chains)
+ (some beta chains)
GCCAG GTTGG C AT
+ (some beta chains)
Beta globin splice mutations: creation of a new acceptor site
NORMAL: INTRON 1 EXON 2
TATTGGTCTATTTTCCCACCCTTAG GCTG 100%
MUTATION:
Normal site used 10% of the time: normal protein from these RNAs
TATT A GTCTATTTTCCCACCCTTAGGCTG
19 nucleotides
TATT A G TCTATTTTCCCACCCTTAGGCTG
New site used 90% of the time: no protein from these RNAs (note the shift in reading frame).
10%
90%
1
0
%
Net result: this allele shows a 90% reduction in β-globin production
A.
Nonsense Mediated Decay
Normal stop codon is downstream or <50 bases upstream from splice junction
Translation
B.
Premature stop codon >50 bases upstream from splice junction
Last exon
Exon/Exon junction
>50 bases mRNA Decay
Last exon
Nonsense Mediated Decay
Origin of premature stop codons
- Improper splicing
- intron retained
- frameshift
- Mutation
Possible consequences of premature stop codons:
- non functional protein
- formation of amyloid
- loss of a regulatory region from a protein that regulates growth
cancer
Nucleus
Nonsense Mediated Decay
Normal Stop Codon
Spliceosomes
Cytoplasm
Protein complexes
(Exon-junction-complexes; EJC)
Exon/Exon junctions
1st round of Translation
Complexes removed by ribosome transit
More translation
Nonsense Mediated Decay
Spliceosomes
Nucleus
Premature stop codon
Normal stop codon
Cytoplasm
Protein complexes
(Exon-junction-complexes; EJC)
Exon/Exon junctions
1st round of Translation mRNA Decay
Complex not removed
Stopped ribosome
Nonsense mediated Decay
Clinical applications (in trials)
• Some drugs that affect the accuracy of codon recognition by ribosomes (such as gentamicin) decrease Nonsense mediated decay.
• Treatment with these drugs allows a low level of expression from genes with premature stop codons.
• Possible treatment for several disorders including some alleles of cystic fibrosis.
Reference: Holbrook et al Nature Genetics 36:801-808 (2004)
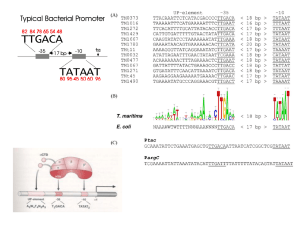
The core promoter
-30 start of transcription +30
TATA box
For most (but not all) promoters, a complex of proteins is assembled around the TATA box, located about 25-30 b.p. upstream from the start site. The consensus sequence of the
TATA box is TATAAA
DNA with TATA box binding protein
Protein
DNA
The core promoter
-30 start of transcription +30
TATA binding protein
The TATA binding protein binds to the TATA box
The core promoter
-30 start of transcription +30
TFII-D
The TATA binding protein is one subunit of a large complex: TFII-D.
TFII-A
The core promoter
TFII-F
-30 start of transcription +30
TFII-B TFII-D
Several other complexes bind to TFII-D.
TFII-A
The core promoter
TFII-F
-30 start of transcription +30
TFII-B TFII-D
RNA pol II
RNA polymerase is recruited to the promoter.
TFII-A
The core promoter
TFII-F
-30 start of transcription +30
TFII-B
TFII-D
TFII-H
RNA pol II
The factor TFII-H plays a key role in initiating transcription by phosphorylating the C-terminal domain of the large subunit of RNA pol II.
CTD: a pol II switch
CTD: The COOH Terminal Domain of the RNA pol II large subunit
…...(Tyr-Ser-Pro-Thr-Ser-Pro-Ser)
52
COOH
CTD: a pol II switch
CTD: The COOH Terminal Domain of the RNA pol II large subunit
…...(Tyr-Ser-Pro-Thr-Ser-Pro-Ser)
52
COOH
ATP
TFIIH
ADP
…...(Tyr-Ser-Pro-Thr-Ser-Pro-Ser)
52
COOH
TFIIH controls the start of transcription
Other kinases
PO
3 phosphorylated Ser 5 of the repeats
ATP
ADP
More phosphorylation of the CTD
CTD: a pol II switch
CTD: The COOH Terminal Domain of the RNA pol II large subunit
Unphosphorylated CTD:
Involved in initiation:
Binding of initiation factors
Phosphorylated CTD:
Involved in elongation & RNA processing
Binds components involved in RNA capping
Binds components involved in RNA splicing
Binds components involved in 3’ end formation
The CTD ties elongation to capping, splicing and 3’-end formation
From Orphanadies & Reinberg (2002) Cell 108:439-51
A model promoter
NR NR
HRE
Fos Jun
TGACTCA
HAT
CREB CREB
GACGTC
Histone Acetyl Transferase
HRE (Hormone
Response Element)
Regulation by hormones such as estrogen which enter the cell
AP-1 (Fos-Jun binding site)
Regulation by growth factors, stress, and various transmembrane signals
CRE (Cyclic AMP
Response Element)
Regulation by cAMP, and by Ca + . Interacts with core promoter (through
CREB Binding Protein) and modifies chromatin structure (through HAT).
Positions of these elements are relatively unimportant
Core Promoter
Binds general transcriptional machinery
SP-1
GGGCGG
TATAAA
SP-1 Site
Provides basal unregulated transcriptional activity. Many genes have multiple
SP-1 sites
Binding of a leucine zipper protein to DNA
Phosphorylation of CREB and the CREB binding protein (CBP)
HAT
Serine 133
Signaling mediated by cAMP and protein kinase A
Hormone
Hormone receptor
Plasma membrane
G
Adenylate cyclase
ATP
G-protein
Inctive
Protein Kinase A cAMP Active
Protein Kinase A
Nuclear membranes
PO
4
PO
4
Active pKA enters the nucleus and phosphorylates
CREB on Serine 133
Core promoter
Phosphorylation of CREB:
- stimulates interactions with several core promoter proteins
- induces binding of HAT and acetylation of histones