Genome, transcriptome and proteome features of malignant cells
advertisement

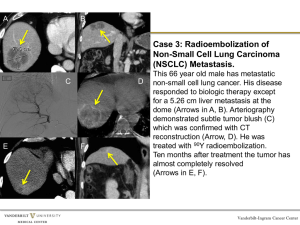
Genome, transcriptome and proteome features of malignant cells as a source of combinatorial biomarkers for clinical translation Ancha Baranova George Mason University, Fairfax, VA Key word to understanding of cancer phenomenon PROGRESSION From bad to worse then to full catastrophe LUNG CARCINOMA PROGRESSION e.g. carcinogen from the smoke e.g. Smoke How long it usually takes: DECADES It takes ≈17 years for a large benign tumor to evolve into an advanced cancer but <2 years for cells within that cancer to acquire the ability to metastasize Sequencing studies show that virtually all of the mutations necessary for metastasis are already present in all of the cells of the antecedent carcinoma. 10.1073/pnas.0712345105; Jones et al., 2008 17 years TRUTH: Majority of human tumors sit in our bodies undiagnosed for decades Major implication for translational research: 1) need for early biomarkers; 2) need for evaluation of how far away the tumor was from sprouting metastasis basis for postsurgical treatment and care Problem: in biomarker research all low-hanging fruits already collected PSA, CEA, AFP…. Single biomarkers have problems with differentiating “grey area” diseases, i.e. inflammatory conditions Solution: Biomarker panels P S A L e v e L Controls Prostate BPH Other Benign cancer cancers genitourinary Barak et al., 1989 diseases Now: from where these biomarker for biomarker panels are usually coming ? A fishing expedition: Differences often reflect Inflammation, fibrosis and other common properties Image courtesy of Purdue University, Dr. W. Andy Tao Two-pronged approach proposed: TUMOR vs. a MIX of various normal cells Perform transcriptome (in silico) and proteome (using antigen screening) subtractions Most TARGETED approach: DISTANCE ANALYSIS: Use entire pattern of gene expression to evaluate of how far away the excised tumor was from sprouting metastasis Most GENERAL approach: Based on previous works of collaborative consortium: Two-pronged approach proposed: TUMOR vs. a MIX of various normal cells Perform transcriptome (in silico) and proteome (using antigen screening) subtractions Most TARGETED approach: Will uncover tumor biomarkers that cannot be masked by antigen production in normal tissue Most Targeted Approach: Consortium Blokhin Russian Oncological Scientific Center, Moscow (sample collection and processing) Caerus Discovery LLC, Manassas, VA (capture of protein biomarker as differentially expressed antigens) Most TARGETED approach: Institute of Chemical Biology and Fundamental Medicine SB RAS, Novosibirsk (development of tumor protein biomarkers as targets for antibody-guided drug delivery) Biomedical Center, StPeterburg (differential display of tumor RNAs) Research Center for Medical Genetics RAMS (non-coding Tumor RNAs) George Mason University, Fairfax, USA (biomarker validation, bioinformatics support and general coordination) Two-pronged approach proposed: DISTANCE ANALYSIS: Use entire pattern of gene expression to evaluate of how far away the excised tumor was from sprouting metastasis Most GENERAL approach: Simple words explanation forusing entire transcriptome as biomarker • Who is that? -- Instant answer (same person, Darwin) Biomarker approach: X: Distance from the end of nose to the upper lip Y: Diameter of the eye orbit Z: Number of hairs in the beard Two-pronged approach proposed: Blokhin Russian Oncological Scientific Center, Moscow (sample collection and processing) Institute of Chemical Biology and Fundamental Medicine SB RAS, Novosibirsk (next gene sequencing and microarray) DISTANCE ANALYSIS: Use entire pattern of gene expression to evaluate of how far away the excised tumor was from sprouting metastasis Vavilov Institute of General Genetics (next gene sequencing and microarray) Research Center for Medical Genetics RAMS (distance analysis of samples based on summed evaluation of non-coding RNAs) Most GENERAL approach: University of Arkansas for Medical Sciences (Little Rock, AK) (distance analysis of whole genome patterns) George Mason University, Fairfax, USA (development of attractor descriptors, bioinformatics support and general coordination) VISION for the FUTURE New generation of tumor markers specific to the malignancy but not to the any normal cell type will allow: 1) True early diagnostics of dormant tumors 2) More specific antibody-guided delivery of therapeutic agents to the tumors VISION for the FUTURE Comparison of prognoses for This tumor and That tumor Same strategy can be realized using NextGen Seq Very possible; low sequence coverage of transcripts will be enough; cost-efficient cut-off needs to be determined VISION for the FUTURE REFERENCE LAB Profiles 100s normal samples for prostate; Establishes its own, equipment-specific NORMAL SPACE For every single tumor sample, the distance from normal tissue space is measured How bad is my tumor? Answer: exact distance from “Ideal” norm Your tumor