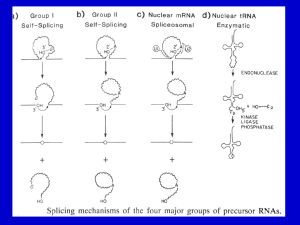
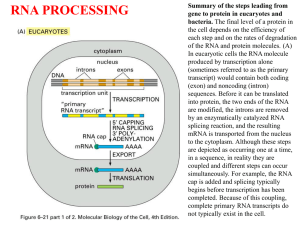
RNA SPLICING AND PROCESSING
CHAPTER 21 (GENES X)
1
Introduction to RNA processing
Transcription elongation is tightly coupled to RNA processing.
Eukaryotic mRNAs are modified at their beginning,
middle and end
Why modify mRNA?
Assess if mRNA is intact
Provides a regulatory mechanism for the amount of protein
produced by a gene
Introns-different forms of a protein from the same gene
2
Introduction to RNA processing
I. mRNA processing (eukaryotes)
1. 5´ capping
2. 3´ cleavage and polyadenylation
3. RNA splicing (nuclear pre-mRNA)
II. Nuclear pore complex overview
III. rRNA and tRNA processing overview
3
mRNA processing
• Occurs in eukaryotes (but a few bacterial
cases exist)
• Primary RNA polymerase II transcripts
(pre-mRNA) are usually not functional
• RNA is processed following (or during)
transcription
4
3 steps of mRNA processing occurs in the nucleus
5’ CAP
Poly-A tail
Splicing
5
Eukaryotic mRNA processing
overview
6
The 5’Cap
7
CAPTHIS IS the 5’ end of
an mRNA!
Marks mRNA for export to
cytoplasm for translation
8
The capping reaction is performed by three enzymes acting
in succession:
A phosphatase: removes one phosphate from the 5’ end
of the nacent RNA,
A guanyl transferase: adds GMP in a reverse linkage (5’
to 5’ instead of 5’ to 3’),
A methyl transferase: adds the methly group to the
guanosine.
9
10
5´ capping
• 5´ end of pre-mRNA
is covalently
modified
• 7-methylguanosine is
added
• Linked 5´ to 5´
• Occurs shortly after
initiation
11
Function of 5´ cap
•
•
•
•
Protection from degradation
Increased translational efficiency
Transport to cytoplasm
Splicing of first exon
12
Do all RNA polymerases CAP?
Pol I and III do not have the CTD, and do not cap
their RNAs
The cap distinguishes mRNAs
13
Structure of two human genes showing the arrangement of exons
and introns.
(A) The relatively small b-globin gene, which encodes one of the
subunits of the oxygen-carrying protein hemoglobin, contains 3 exons
(see also Figure 4–7). (B) The much larger Factor VIII gene contains
26 exons; it codes for a protein (Factor VIII) that functions
14
Variation in intron and exon
lengths in the human,
worm, and fly genomes.
(A) Size distribution of exons.
(B) Size distribution of introns.
Note that exon length is
much more uniform than
intron length.
15
Splicing
16
Introduction to RNA processing
I. mRNA processing (eukaryotes)
3. RNA splicing (nuclear pre-mRNA)
A. Introns, exons, and splicing
B. Spliceosome and snRNAs
C. Self-splicing
D. Alternative splicing
17
RNA Splicing
Primary transcripts (in eukaryotes) are sometimes “spliced” to
remove non-coding regions “introns” from coding regions “exons”
The exon regions are spliced together to form the mature mRNA
hnRNA
addition of cap, polyA tail
- Poly A Tail
5’ Cap-
Splicing
- Poly A Tail
5’ Cap-
Mature mRNA
18
Types of mRNA Splicing
Types I & II: self-splicing of catalytic RNA
sequences (Ribozymes)
- Two types reflect different ribozyme types
Type III: occurs in a protein - RNA complex
- Responsible for nearly all splicing
- It has been speculated that the RNA component of this
structure is the catalytic component which would also
make it a ribozyme
19
Prevalence Of Splicing
Type III:
Eukaryotes - most mRNAs in vertebrates
- many mRNAs in invertebrates
no type III mRNA splicing
- some mRNAs in unicellular
in Prokaryotes!
eukaryotes
mRNA:
Type I & II:
Eukaryotes - rRNA in tetrahymena nuclei
(Type I)
- mRNA & rRNA in fungal
mitochondria (I & II)
- mRNA in some chloroplasts (II)
20
Prokaryotes - mRNA in bacteriophage T4 (I)
Mechanism of pre-mRNA Splicing
(Spliceosome Mediated) Type III
- Introns have conserved sequences at the splice junctions
- SnRNPs (Small nuclear ribonuclear proteins) binds critical
sites on the pre-mRNA
- pronounced ‘SNURPs’
- these are complexes containing both protein and small
RNAs
- the small RNAs are transcribed by RNA polymerase III
- they then associate with accessory proteins
- the complex then recognizes critical sites for splicing by base
21
pairing
22
23
The consensus nucleotide sequences in an RNA molecule that signal the beginning and the end of most
introns in humans. Only the three blocks of nucleotide sequences shown are required to remove an intron
sequence; the rest of the intron can be occupied by any nucleotide. Here A, G, U, and C are the standard RNA
nucleotides; R stands for either A or G; Y stands for either C or U. The A highlighted in red forms the
branch point of the lariat produced by splicing. Only the GU at the start of the intron and the AG at its end
are invariant nucleotides in the splicing consensus sequences. The remaining positions (even the branch point
A) can be occupied by a variety of nucleotides, although the indicated nucleotides are preferred. The
distances along the RNA between the three splicing consensus sequences are highly variable; however, the
distance between the branch point and 3' splice junction is typically much shorter than that between the 5'
24
splice junction and the branch point.
- consensus sequences are conserved throughout eukaryotes
Conservation of sequence is expected with recognition of sequences
being done by base pairing with snRNP’s RNA component
25
26
The RNA splicing reaction.
(A) In the first step, a specific adenine
nucleotide in the intron sequence (indicated
in red) attacks the 5' splice site and cuts the
sugar-phosphate backbone of the RNA at
this point. The cut 5' end of the intron
becomes covalently linked to the adenine
nucleotide, as shown in detail in (B),
thereby creating a loop in the RNA
molecule. thereby creating a loop in the
RNA molecule. The released free 3'-OH end
of the exon sequence then reacts with the
start of the next exon sequence, joining the
two exons together and releasing the intron
sequence in the shape of a lariat. The two
exon sequences thereby become joined into
a continuous coding sequence; the released
intron sequence is degraded in due course.
27
28
Excised intron sequence in
form of a lariant structure
29
Nuclear pre-mRNA splicing
• Introns: non-coding sequences
• Exons: coding sequences
• RNA splicing: removal of introns and
joining of exons
• Splicing mechanism must be precise to
maintain open reading frame
• Catalyzed by spliceosome (RNA + protein)
30
Classes of intronic splicing: splicesome
• Major pathway for splicing and is used for processing
of all pre-mRNAs in eukaryotes
• Splicesome is a complex of five small nuclear
ribonucleoprotein particles (snRNPs).
• Each snRNPs consists of a small nuclear RNAs
(snRNA) associated specifically with proteins.
• Large, ~60S (size of a ribosome), 5 snRNAs (U1, U2,
U4, U5, U6) and roughly 50 proteins!
• Splice at specific markers with the mRNA using the
31
snRNA for recognition and catalysis!
Once all of the different snRNPs associate with their
appropriate targets on the pre-mRNA
the entire (very large) complex is called a:
Spliceosome
32
Splicesomal mechanism
•
•
snRNA U1 has sequences that are complementary to 5’ splice site
– U1 plus its protein component is the U1 snRNP
upon U1 snRNP binding, U2, U4, U5 and U6 snRNP assemble
– ATP is required for assembly but not catalysis! (needed for RNA helicases that
allow alternative pairing schemes)
Proteins removed from this view for clarity!
33
Secondary structure model of human U1 snRNP. The region where
it recognizes the pre-mRNA is also shown
34
• The E complex contains:
– U1 snRNP bound at the 5′ splice
site
– the protein U2AF bound to a
pyrimidine tract between the
branch site and the 3′ splice site
– SR proteins connecting U1
snRNP to U2AF
Important point is recognition
and complementarity of 5’
splice site sequences with U1
RNA sequences.
35 35
Figure 26.10
Splicesomal
mechanism
ATP is required for assembly but not for catalysis
36
The RNA splicing mechanism.
RNA splicing is catalyzed by an
assembly of snRNPs (shown as colored
circles) plus other proteins (most of
which are not shown), which together
constitute the spliceosome. The
spliceosome recognizes the splicing
signals on a pre-mRNA molecule,
brings the two ends of the intron
together, and provides the enzymatic
activity for the two reaction steps (see
Figure 6–26). The branch-point site is
first recognized by the BBP (branchpoint binding protein) and U2AF, a
helper protein. In the next steps, the U2
snRNP displaces BBP and U2AF and
forms base pairs with the branch- point
site consensus sequence, and the U1
snRNP forms base-pairs with the 5'
splice junction (see Figure 6–30). At
this point, the U4/U6∑U5 “triple”
snRNP enters the spliceosome. 37
In this triple snRNP, the U4 and
U6 snRNAs are held firmly
together by base-pair interactions
and the U5 snRNP is more loosely
associated. Several RNA–RNA
rearrangements then occur that
break apart the U4/U6 base pairs
(as shown, the U4 snRNP is
ejected from the splicesome
before splicing is complete) and
allow the U6 snRNP to displace
U1 at the 5' splice junction (see
Figure 6–30). Subsequent
rearrangements create the active
site of the spliceosome and
position the appropriate portions
of the pre-mRNA substrate for the
splicing reaction to occur.
Although not shown in the figure,
each splicing event requires
additional proteins, some of which
hydrolyze ATP and promote the
38
RNA–RNA rearrangements.
Doublechecking of intron boundary sequences
Several of the rearrangements that take place in the spliceosome during pre-mRNA splicing.
Shown here are the details for the yeast Saccharomyces cerevisiae, in which the nucleotide sequences
involved are slightly different from those in human cells. (A) The exchange of U1 snRNP for U6 snRNP
occurs before the first phosphoryl-transfer reaction (see Figure 6–29). This exchange allows the 5' splice site
to be read by two different snRNPs, thereby increasing the accuracy of 5' splice site selection by the
spliceosome. (B) The branch-point site is first recognized by BBP and subsequently by U2 snRNP; as in (A),
this “check and recheck” strategy provides increased accuracy of site selection. The binding of U2 to the
branch-point forces the appropriate adenine (in red) to be unpaired and thereby activates it for the attack on
the 5' splice site (see Figure 6–29). This, in combination with recognition by BBP, is the way in which the
39
spliceosome accurately chooses the adenine that is ultimately to form the branch point.
40
U5 is present here
(C) After the first phosphoryl-transfer reaction
(left) has occurred, U5 snRNP undergoes a
rearrangement that brings the two exons into close
proximity for the second phosphoryl-transfer
reaction (right). The snRNAs both position the
reactants and provide (either all or in part) the
catalytic site for the two reactions. The U5 snRNP
is present in the spliceosome before this
rearrangement occurs; for clarity it has been
omitted from the left panel. All of the RNA–RNA
rearrangements shown in this figure (as well as
others that occur in the spliceosome but are not
shown) require the participation of additional
proteins and ATP hydrolysis.
U5 snRNP undergoes
rearrangement
41
The E complex forms by interactions involving both splice sites.
The commitment (E) complex
forms by successive addition of
U1 SnRNP to the 5’ splice site,
U2AF to the pyrimidine track/3’
splice site, and the bridging
protein SF1/BBP.
• ASF/SF2 (a general splicing factor in the SR class)
• U2AF splicing factor (member of SR, arg/ser rich proteins)
U2AF65 contacts pyrimidine track
U2AF35 contacts dinucleotide AG, 3’ splice site.
• SF1 splicing factor connects U2AF to U1 snRNP bound to 5’ splice site.
42
Introns ends can be recognized by either of two pathways.
Intron definition
Exon definition
43
A splicesome forms via several complex.
E complex: Formation of commitent
Complex in which U1 is basepaired with the
5’ splice site
A complex: U2 addition to basepair with the
branch site in the presence of ATP
B1 complex: Joining of U4.6/U5 tri-snRNPs
B2 complex: U1 and U4 release
Formation of the catalytic center in which U6
basepairs with U2;U2 reamins basepaired
with the branch site; U5 interacts with both
exons through its loop.
C1 complex: The first step of
transesterification
5’ splice site cleaved, lariant formed
C2 complex: The second step of
transesterification
3’ splice site cleaved, exaons ligated
44
The exon definition hypothesis. According to one proposal, SR proteins bind to each exon sequence in
the pre-mRNA and thereby help to guide the snRNPs to the proper intron/exon boundaries. This
demarcation of exons by the SR proteins occurs co-transcriptionally, beginning at the CBC (cap-binding
complex) at the 5' end. As indicated, the intron sequences in the pre- mRNA, which can be extremely long,
are packaged into hnRNP (heterogeneous nuclear ribonucleoprotein) complexes that compact them into
more manageable structures and perhaps mask cryptic splice sites. Each hnRNP complex forms a particle
approximately twice the diameter of a nucleosome, and the core is composed of a set of at least eight
different proteins. It has been proposed that hnRNP proteins preferentially associate with intron sequences
and that this preference also helps the spliceosome distinguish introns from exons. However, as shown, at
least some hnRNP proteins may bind to exon sequences but their role, if any, in exon definition has 45
yet to
be established.
3´ cleavage and polyadenylation
• RNA polymerase II does not usually
terminate at distinct site
• Pre-mRNA is cleaved ~20 nucleotides
downstream of polyadenylation signal
(AAUAAA)
• ~200 AMPs are then added to the 3´ end
• Almost all mRNAs have poly(A) tail
46
• The sequence
AAUAAA is a signal
for cleavage to
generate a 3′ end of
mRNA that is
polyadenylated.
• The reaction requires
a protein complex
that contains:
– a specificity factor
– an endonuclease
– poly(A) polymerase
The 3’ end of mRNA is generated by cleavage. mRNA is stabilized by
polyadenylation.
• The specificity factor and endonuclease cleave RNA
downstream of AAUAAA.
• The specificity factor and poly(A) polymerase add ∼200 A
residues processively to the 3′ end.
• A-U-rich sequences in the 3’ tail control cytoplasmic
polyadenylation or deadenylation during Xenopus embryonic
development.
48
Consensus nucleotide sequences that direct cleavage and polyadenylation to form the 3' end of a
eucaryotic mRNA.
These sequences are encoded in the genome and are recognized by specific proteins after they are
transcribed into RNA. The hexamer AAUAAA is bound by CPSF, the GU-rich element beyond the
cleavage site by CstF (see Figure 6–38), and the CA sequence by a third factor required for the cleavage
step. Like other consensus nucleotide sequences discussed in this chapter, the sequences shown in the
49
figure represent a variety of individual cleavage and polyadenylation signals.
Basic steps of poly-Adenylation
•
•
Most eukaryotic mRNA have 80-250 Ade added to their 3’ end by a multi-step process.
CTD of Pol II has a 7aa (x52) that strongly stimulates polyadenylation
– Endonuclease (complex) cleaves 10-30 nt downstream of AUAAAA
– PABPI recruits polyA pol
– Poly-A pol adds A in association with PABPII
50
3’ modification-addition of a poly A tail
CSTF- cleavage stimulation factor
CPSF- cleavage and polyadenylation specificity factor
PAP-
PolyA polymerase
PABP- Poly A binding proteins
51
3’ modification-addition of a poly A tail
CSTF- cleavage stimulation factor
CPSF- cleavage and polyadenylation specificity factor
PAP-
PolyA polymerase
PABP- Poly A binding proteins
52
Some of the major steps in generating the 3' end of a eukaryotic mRNA.
This process is much more complicated than the analogous process in bacteria, where the RNA polymerase simply
stops at a termination signal and releases both the 3' end of its transcript and the DNA template.
53
Function of poly(A) tail
• Increased mRNA stability
Protects mRNA
In cytoplasm poly(A) size decreases due to RNAses, however poly(A) polymerase
continues rebuilding: No poly(A) tail…destroyed!
• Increased translational efficiency
Necessary for translation (site of binding by poly(A)-binding protein I (PABI)
Recruits mRNA to polylysomes so that translation can be initiated
• Splicing of the last intron
54
Degradation of RNA
• mRNAs are degraded at different rate
(control of gene expression)
• Life time varies from several seconds to
several generations. T1/2= 3hours for
vertebrate and 1.5minutes for bacteria.
• RNA is degraded from 5’ to 3’.
• 3’-sequence often inhibit 3’-5’
exoribonuleases.
55
Are you ready for export?
Cap
pA tail
No snRNPs bound
Other proteins bind that
Indicate mRNA is ready
56
Nuclear transport
• Nucleus houses DNA and is site of
transcription and RNA processing
• Cytoplasm is site of translation
• RNAs must leave nucleus
• Some proteins must enter nucleus
• Nucleus is separated from cytoplasm by
nuclear envelope (double biomembrane)
• Transport across nuclear envelope is
through numerous nuclear pores
57
Nuclear pore complex
• Portal through
nuclear
envelope
• Mediates
traffic in and
out of nucleus
• Very large
multiprotein
complex
58
THE NUCLEAR PORE COMPLEX (NPC) I
1. 125 x 106 D, spans the inner and outer membranes of the NE
2. Octagonal symmetry
59
3. Represents the transit path for molecules going into and out of the nucleus
Transport of a large mRNA molecule
through the nuclear pore complex.
(A) The maturation of a Balbiani Ring
mRNA molecule as it is synthesized by
RNA polymerase and packaged by a
variety of nuclear proteins. This drawing
of unusually abundant RNA produced by
an insect cell is based on EM
micrographs such as that shown in (B).
60
The EJC (Exon Junction Complex) binds to RNA by recognizing the splicing complex.
61
Transcript export
Proteins associated with mRNA mark it for export
Only mature mRNA is exported from nucleus
Exit via nuclear pore complexes
62
A REF protein binds to a splicing factor and remains with the spliced
RNA product. REF binds to an export factor that binds to the nuclear
pore.
63
Schematic illustration of an “export- ready” mRNA molecule and its transport through the
nuclear pore. As indicated, some proteins travel with the mRNA as it moves through the pore,
whereas others remain in the nucleus. Once in the cytoplasm, the mRNA continues to shed
previously bound proteins and acquire new ones; these substitutions affect the subsequent
translation of the message. Because some are transported with the RNA, the proteins that become
bound to an mRNA in the nucleus can influence its subsequent stability and translation in the
cytosol. RNA export factors, shown in the nucleus, play an active role in transporting the mRNA to
the cytosol (see Figure 12–16). Some are deposited at exon-exon boundaries as splicing is
completed, thus signifying those regions of the RNA that have been properly spliced.
64
Putting it all together
1.
2.
3.
4.
5.
6.
7.
8.
9.
Get at gene through chromatin
Pol II recruited to bind promoter via a host of TFs (up and down)
Rearrangement of PIC to start elongation
Capping (cotranscriptionally!)
Splicing (+ alt splicing) (cotranscriptionally!)
Termination polyadenylation
Export to cytoplasm
Translation
turnover
All of these steps are regulated and all can contribute to increasing
or decreasing the levels of mature mRNA!
65
ALTERNATIVE SPLICING MECHANISMS
66
ALTERNATIVE SPLICING
A COMMON PRE-mRNA IS SPLICED INTO MULTIPLE
mRNA ISOFORMS DIFFERING IN THEIR COMBINATION
OF EXON SEQUENCES.
THIS GENERATION OF MULTIPLE mRNAS FROM A
SINGLE GENE PARTIALLY UNDERLIES THE APPARENT
DISCREPANCY BETWEEN GENE NUMBER AND THE
COMPLEXITY OF THE ORGANISM.
67
Alternate Splicing
Splicing at alternate splice sites can lead to the production of
several gene products from one gene
- Proteins involved in splicing can bring alternate domains of
pre-mRNA together to give alternate splicing events
- Alternate splicing can be regulated
- differential splicing can occur at different times during development
and in different cell types
GENE
(hnRNA)
2
1
1
2
3
3
4
4
2 Different
mRNA products
1
3
4
68
69
70
71
• The regulation of RNA splicing can
generate different versions of protein in
different cell types, according to the needs
of cell.
• Tropomyosin for example is produced in
specialized forms in different types of cells.
72
Example of alternate splicing with rat -tropomyosin
Alternative splicing of the a-tropomyosin gene from rat. a-tropomyosin is a coiled-coil protein that
regulates contraction in muscle cells. The primary transcript can be spliced in different ways, as indicated in
the figure, to produce distinct mRNAs, which then give rise to variant proteins. Some of the splicing patterns
are specific for certain types of cells. For example, the a- tropomyosin made in striated muscle is different
73
from that made from the same gene in smooth muscle. The arrowheads in the top part of the figure demark
the sites where cleavage and poly-A addition can occur.
Alternative splicing!
• ~30% of human genes are subject to alternative
splicing
• Regulated as a function of cell/developmental
stage as well as in response to signals
• Range of exons from 1 to 54 (HGP)
– Percent of human DNA in exons 3%
– Total number of genes ~33,000 but alternative
splicing increases the diversity of protein
74
Alternative Splicing Patterns
ALTERNATIVE 5’ SPLICE SITES
ALTERNATIVE 3’ SPLICE SITES
SINGLE ALTERNATIVE EXON:
EITHER INCLUDED OR SKIPPED
MULTIPLE ALTERNATIVE EXONS:
ONE OR OTHER CAN BE CHOSEN
INTRON RETAINED AND TRANSLATED
From BR Graveley, Trends in Genomics:
17(2001)100-107
75
Four patterns of alternative splicing: In each case single type of RNA transcript is spliced in two
alternative ways to produce two distinct mRNAs (1 and 2). The dardk blue boxes mark on exon
sequences that are retained in both mRNAs. The light blue boxes mark possible exon sequences that are
included in only one of the mRNAs. The boxes are joined by red lines to indicate where intron
sequences (yellow) are removed.
76
Negative and positive control of alternative RNA splicing. (a) Negative control, in which a
repressor protein binds to the primary RNA transcript in tissue 2, thereby preventing the
splicing machinery from removing an intron sequence. (B) positive control, in which the
splicing machinery is unable to efficiently remove a particular intron sequence without
assistance from an activator protein.
77
Alternative Splicing can change a
protein’s properties
78
Sex determination in Drosophila
• Individuals with an X/A ratio of 1 (normally
two X chromosomes and two sets of
autosomes) develop as females.
• X/A ratio of 0.5 (normally one X
chromosomes and two sets of autosomes)
develop as males.
• This ratio is assigned early in development
and is remembered thereafter by each cell.
79
Sex determination in Drosophila
Three crucial gene products
transmit information about this
ratio to the many other genes that
specify male and female
characteristics.
5th edition, Chapter 7: control of gene expression, page 479-483
80
A cascade of changes in gene expression that determines the sex of a fly through alternative splicing.
81
SEX DETERMINATION IN DROSOPHILA: A
CASCADE OF ALTERNATIVE SPLICING
82
SECOND STEP OF SPLICING OF SXL PRE-MRNA
SPF45=SECOND STEP SPLICING FACTOR
FUNCTIONS
IN MALES
SXL BINDS UPSTREAM OF AG,
THEREBY INTERACTING WITH
AND INHIBITING SPF45
83
ALTERNATIVE SPLICING OF TRA PRE-MRNA
SXL binds to the pyrimidine-rich stretch of nucleotides that is part of the standard
splicing consensus sequence and blocks access by normal splicing factor, U2AF.
TRUNCATED
TRA PROTEIN
FULL-LENGTH
TRA PROTEIN
84
Tra binds to specific RNA sequence in an exon and with Tra2 activates a
normally suboptimal signal by the binding of U2AF. Tra is a positive activator
of RNA splicing.
ONLY PRODUCED
IN FEMALES
SR protein
TRANSCRIPTIONAL REPRESSOR
OF MALE-SPECIFIC GENES
SR protein
MALE
TRANSCRIPTIONAL REPRESSOR OF FEMALE-SPECIFIC GENES
85
Here is what an alternative splice
might look like on a protein
Splice product 1
Splice product 2
• Protein may have different ligand specificity or
enzyme activity etc…
86
Alternative splicing of Slo gene
• The mammalian cochlea is a snail-like structure of the
inner ear that contains hair cells organized along a basilar
membrane.
• There are four rows of hair cells, one inner hair cell and
three outer hair cells.
• The hair cells are turned to unique narrow sound
frequencies along the basilar membrane and creating a
tonic gradient.
• At one end the hair cells are tuned to respond to a
frequency of 20 Hz, other side cells respond to 20,000 Hz.
87
Alternative splicing of Slo gene
• Hair cells tunning is thought to be mediated, in part, by the alternative
splicing of transcripts expressed from the calcium-activated potassium
channel gene, Slo.
• Isoforms of the Slo gene lacking sequences encoded by the STREXexon have fast deactivation kinetics and low ca++ sensitivity,
• where as STREX containing sequences have a slower deactivation
kinetics and higher ca++ sensitivity.
88
Alternative splicing in human slo gene; >500 possible isoforms.
(believed to be involved in hearing sounds of different frequencies.)
From BR Graveley, Trends in Genetics:17(2001)100-107.
Alternative splicing, alternative exons are grey boxes
Constitutive splicing
Isoform of Slo protein lacking sequences encoded by STREX exon
89
Neuronal signaling and
synaptogenesis
• Neurexins are family of a neuronal proteins present in
vertebrates that have important functions as receptors for
neuropeptides and adhesion molecules that participate in
synaptogenesis.
• b-neurexins containing exon 20-encoded sequences can
inhibit synaptogenesis, whereas exon-20 containing
neurexin does not.
The relative ratio of the two protein isoforms could determine
whether functional synapsis are formed or broken.
90
Alternative splicing in human neurexin 1 gene; >300 possible isoforms.
(believed to be involved in making and breaking synaptic connections between
neurons.) From BR Graveley, Trends in Genetics:17(2001)100-107.
The LNS domains of neurexin and agrin undergo alternative splicing that modulates their
affinity for protein ligands in a neuron-specific manner.
91
ALTERNATIVE SPLICING IN THE NERVOUS
SYSTEM:
GENERATION OF MUCH OF THE ENORMOUS DIVERSITY
NEEDED IN THE PROTEINS INVOLVED IN FORMING
SPECIFIC SYNAPTIC CONNECTIONS AND IN MEDIATING
SYNAPTIC TRANSMISSION
92
Alternative splicing of Drosophila DSCAM gene: DSCAM proteins are axon guidiance receptors that help to
direct growrh cones to their appropriate targets in the developing nervous system. The final mRNA contains 24
exons, four of which (denoted A, B, C and D) are present in DCAM gene as arrays of alternative exons. Each
RNA contains 1 of 48 alternative exon A (red), 1 of 48 alternative exon of B (green), 1 of 33 alternative exon of
C (yellow). If all possible splicing combinations are used, 38,016 different proteins could be in principle be
produced from the gene. Each variant DSCAM protein would fold roughly the same structure. (predominantly
a series of extracellular immunoglobulin like domains linked to a mambrane-spaining region) but the amino
acid sequence of the domains would vary according to the splicing pattern. It is possible that this receptor
deversity contributes to the formation of neural circuits, but the precise properties and functions of the many
DSCAM variants are not yet understood.
93
DROSOPHILA DSCAM GENE ENCODES AN AXON GUIDANCE RECEPTOR
THAT CAN EXPRESS 38,016 mRNAS VIA ALTERNATIVE SPLICING
EXON 4 ALTERNATIVE SPLICING IS DEVELOPMENTALLY REGULATED:
EXON 4.2 INFREQUENTLY USED IN EARLY EMBRYOS, PREDOMINANT IN
ADULTS
94
Alternative splicing in Dorsophilia Dscam gene; 38,000 possible isoforms.
(believed to be involved in Axon migration and connection.)
From BR Graveley, Trends in Genetics:17 (2001)100-107.
Neurons expressing the form of Dscam shown on the right will be attracted in a
different direction than neurons expressing the form on the left.
95
TISSUE-SPECIFIC RNA SPLICING FACTOR IN THE NERVOUS SYSTEM: NOVA-1, A
REGULATOR OF ALTERNATIVE SPLICING IN THE BRAINSTEM AND SPINAL CORD
Nova-1 null mice show specific splicing defects in two inhibitory receptor pre-mRNAs, glycine
alpha2 exon 3A (GlyRalpha2 E3A) and GABA(A) exon gamma2L. Nova protein in brain
extracts specifically bound to a previously identified GlyRalpha2 intronic (UCAUY)3 Nova
target sequence, and Nova-1 acted directly on this element to increase E3A splicing in
cotransfection assays (Neuron. 2000 Feb;25(2):359-71).
96
Regulation of the sites of RNA cleavage and poly-A addition determines whether an antibody molecule is secreted or remains membranebound. In stimulated B lymphocytes(left), a long RNA transcript is produced, and the intron sequence near its 3’ end is removed by
RNA splicing to give rise to an mRNA molecule that codes for a membrane bound antibody molecule. In contrast, after antigen
stimulation (right) the primary RNA transcript is cleaved upstream from the spliced site in front of the last exon sequence. As a result,
some of the intron sequence that is removed from the long transcript remains as coding sequence in the short transcript. These are the
97
nucleotide sequences that encode the hydrophilic C-terminal portion of the secreted antibody molecule.
Self-splicing introns
• Introns that splice themselves out of genes
• Ribozymes: catalytic RNA
• Two major types (group I and group II)
98
Self-Splicing (Type I)
T.R. Cech, ~1982 made a revolutionary discovery:
RNA could have catalytic activity
RNA enzyme = ‘Ribozyme’
- For this type of splicing, only require:
1. Pre-rRNA
2. Mg2+
3. G (GDP,GMP, guanosine) –guanine does not
work
99
Self-Splicing Introns fold so as to:
1. Bring 5’ & 3’ splice sites together
2. Form a G-binding pockets close to the splice sites
3. Mg2+ :
1. Stabilizes the folded RNA structure
2. Undoubtedly helps to lower G for bond transfer
3’-OH of the {G} cofactor initiates the reaction
- attacks phosphate at the 5’ splice site.
- itself becomes attached to the 5’ end of the intron
- Role equivalent to branch site adenine in type III splicing
100
Classes of intronic splicing: Group I
Found in some mitochondrial/chloroplast rRNAs, tRNAs and mRNAs
• Requires guanosine as cofactor
(not energy)
• 3’ OH acts as nucleophile in
the two transesterification
reactions
– G attacks 5’ splice site to
form 3’-5’ phosphodiester
(to 5’ end of intron)
– 3’ OH of displaced exon
acts as a nucleophile and
attacks the 3’ end of of the
intron , yields precise
excision of intron!
101
Group II self-splicing introns
• Found in plant and fungal organelles
• Mobile DNA elements (nonviral
retrotransposons)
• Encode maturases (proteins) needed for
physiological splicing
• Similar secondary structure (and splicing
mechanism) as snRNAs in spliceosome
102
Classes of intronic splicing: Group II
often in mitochondrial/chloroplast genes in fungi, algae and plants
• Similar to Group I except
nucleophile in first
transesterification step is 2’ OH
of an intronic A
• Forms a “lariat” intermediate
from 2’-5’ bond
• again, no energy required
• Like Group I splicing it is entirely
dependent on a complex 3D fold
of the RNA for activity
103
Predicted secondary structures
104
105
106
107