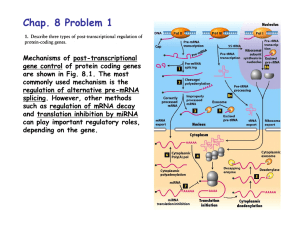
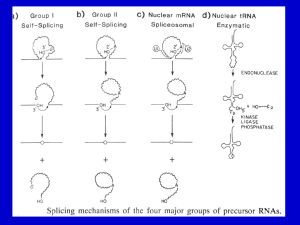
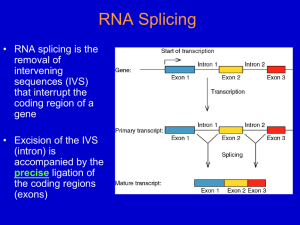
Discovery of Alternative Splicing
First discovered with an Immunoglobulin heavy
chain gene (D. Baltimore et al.)
•
Alternative splicing gives two forms of the protein
with different C-termini
– 1 form is shorter and secreted
– Other stays anchored in the plasma membrane
via C-terminus
~40% of human genes produce alternatively spliced
transcripts!
Alternative splicing of the mouse
immunoglobulin μ heavy chain gene
S-signal peptide
V- variable region
Red- untranslated reg.
Fig. 14.38
C - constant region
green – membrane anchor
yellow – end of coding reg. for secreted form
Regulation of Alternative splicing
•
Sex determination in Drosophila involves 3 regulatory
genes that are differentially spliced in females versus
males; 2 of them affect alternative splicing
1.
Sxl (sex-lethal) - promotes alternative splicing of tra
(exon 2 is skipped) and of its own (exon 3 is skipped)
pre-mRNA
Tra – promotes alternative splicing of dsx (last 2 exons
are excluded)
Dsx (double-sex) - Alternatively spliced form of dsx
needed to maintain female state
2.
3.
Fig. 14.38
Alternative
splicing
in Drosophila maintains the female state.
Alternative
splicing
Sxl and Tra are SR proteins!
Tra and Tra-2 bind a repeated element in exon 4 of dsx mRNA,
causing it to be retained in mature mRNA.
Fig. 14.39
Trans-Splicing (Ch. 16.3)
• Intermolecular splicing of pre-mRNAs
• First discovered in African trypanosomes, a
disease(African Sleeping Sickness)-causing
parasitic protozoan.
• The mRNAs had 35 nt not encoded in the main
gene – called the spliced leader sequence.
• Spliced leader (SL) is encoded separately, and
there about 200 copies in the genome .
• SL primary transcript contains ~100 nt that
resemble the 5’ end of a NmRNA intron.
Organisms that trans-splice nuclear genes.
from Fig. 16.8
Trypanosome
Schistosoma
Ascaris
Trans-splicing also occurs in plant chloroplast and
mitochondrial genes!
Euglena
2 possible models to explain the joining of the
SL to the coding region of a mRNA
1. Primed transcription by SL
2. Trans-splicing model
Fig. 16.9
Trans-splicing in Trypanosomes
SL
Trans-splicing should yield some unique “Y –shaped”
intron-exon intermediates containing the SL half-intron.
Fig. 16.10
Release of the SL half-intron from larger
RNAs by a debranching enzyme.
This result is consistent with a transsplicing model rather than a cis-splicing
mechanism.
Figs. 16.11, 16.12
Some of these organisms (Trypanosomes
and Euglena) also have polycistronic
genes.
Trypanosome
Schistosoma
Ascaris
Euglena
Parasitic Worms
Fig. 16.8
Cap stimulates splicing of the first
intron in a multi-intron pre-mRNA
32P-labeled
substrate
RNAs were
incubated
in a Hela
nuclear
extract.
May have
been
methylation
of Cap in
extract.
Fig. 15.30
Splicing of 1st
intron very
poor with
uncapped premRNA.
CAP Binding Complex (CBP)
• Contains 2 proteins of 80 (CBP80) and
20 (CBP20) kiloDaltons
• Depletion of CBP from a splicing extract
using antibody against CBP80
inhibited splicing of the first intron in a
model pre-mRNA
– Further analysis showed an inhibition of
spliceosome formation
• CBP may be important for spliceosome
formation in vivo on first intron
Poly A-Dependent Splicing of the Last
Intron in a 2-intron pre-RNA
Double-spliced mRNA
Splicing of the 2nd intron in this pre-mRNA is reduced by a mutation
in the polyadenylation signal (wild-type hexamer=AAUAAA).
Splicing of the 1st intron is normal.
Fig. 15.31
RNA Splicing and Disease
• ~ 15 % of the mutations that cause genetic diseases
affect pre-mRNA splicing.
• Many are cis-acting mutations at the splice-sites, the
branch point, or sequences that promote (enhancers)
or inhibit (silencers) splicing of certain exons.
• OMIM (Online Mendelian Inheritance in Man) database of human genetic mutations and disorders
at NCBI, a.k.a. the National Center for Biotechnology
Information) (link on Blackboard)