Proximal promoter
advertisement

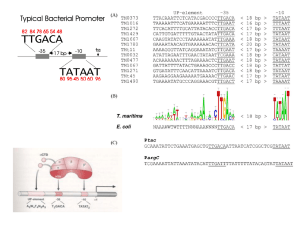
Inferring Transcriptional Regulation Using Transctiptomics Carsten O. Daub September 1st, 2014 StratCan Summer School 2014 Vår Gård, Saltsjöbaden Overview – Levels of Regulation • Genome – SNP – DNA modifications (e.g. methylation) – structural alterations (e.g. genomic rearrangements) • Transcriptome – – – – – – Transcription factors, enhancers/ insulators Promoter RNA splicing miRNA Posttranscriptional modifications (e.g. RNA editing) 3D structure of the genome • Protein – Translation – Posttranslational modifications • Metabolites Central Dogma of Molecular Biology DNA Transcription RNA Translation Protein Francis Crick, 1958 Non coding RNA What is the transcriptome? • The ensemble of all expressed RNA • Protein coding genes • Non-protein coding genes How is the Transcriptome regulated? • Via Promoter – Transcription factors – enhancers – insulators • RNA splicing • miRNA • Posttranscriptional modifications (e.g. RNA editing) • 3D structure of the genome Regulation via the Promoter Transcription • The principle: DNA is copied into RNA by the RNA polymerase (Pol) 5’ Pol 3’ • Transcription initiation is more complex in eukaryotes than in prokaryotes • In eukaryotes several different factors are necessary for the transcription of an RNA polymerase II promoter. http://en.wikipedia.org/wiki/Gene • Initiation – Promoter clearance – Pol2 stalling • Elongation • Termination Figures from http://en.wikipedia.org/wiki/Transcription_(genetics) Transcription Model 5’ Pol 3’ Transcription Pre-mRNA (precursor) Capping ( ) Splicing Polyadenylation mRNA AAAAAAAAAAA Transcription Factor (TF) Binding • TFs bind to specific sites in the DNA • Sets of TFs can function as cisregulatory modules (CRM) Nature Reviews Genetics 5, 276-287 (April 2004) Specific TF Binding • Transcription factors bind to specific DNA sequences • Databases of TF binding sequence motifs – JASPAR, TRANSFAC IRF8 binding motif DNA IRF8 Promoter Region Transcription start site (TSS) Distal promoter [-10k, -250] Proximal promoter [-250, -34] Core promoter [-34, -1] Promoter Region • Core promoter – the minimal portion of the promoter required to properly initiate transcription – – – – Transcription Start Site (TSS) Approximately -34 A binding site for RNA polymerase General transcription factor binding sites • Proximal promoter – the proximal sequence upstream of the gene that tends to contain primary regulatory elements – Approximately -250 – Specific transcription factor binding sites • Distal promoter – the distal sequence upstream of the gene that may contain additional regulatory elements, often with a weaker influence than the proximal promoter – Anything further upstream (but not an enhancer or other regulatory region whose influence is positional/orientation independent) – Specific transcription factor binding sites Transcription in eukaryotes • In eukaryotes, several different factors are necessary for the transcription of an RNA polymerase II promoter. Name Location RNA transcribed RNA Polymerase I nucleolus ribosomal RNA (rRNA) RNA Polymerase II nucleus messenger RNA (mRNA) and most small nuclear RNAs (snRNAs) RNA Polymerase III nucleus transfer RNA (tRNA) and other small RNAs Identifying the TF regulators • How much is a TF binding site used – Observed expression of all genes – Predicted site count • Motif Activity Response Analysis (MARA) FANTOM4 – A Systems Approach Monoblast-like THP-1 cells were stimulated by PMA to differentiate them into monocyte-like cells. 10 time point samples were collected during differentiation. Monocyte-like Monoblast-like 0 1 2 4 6 12 24 48 72 96 hour PMA Replicates Microarray check Deep CAGE RIKEN1 RIKEN3 RIKEN5 RIKEN6 TF qRT-PCR Not good Illumina (47K probes) 10 time points miRNA microarray Cap Analysis of Gene Expression (CAGE) CAGE library preparation CAGE data digital processing Sequencing Figure based on [1] Tag cluster (TC) 1 Carninci, P. et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nature genetics 38, 626–35 (2006) CAGE identifies the active set of promoters Alternative promoter usage for PTPN6 HeLa Promoter THP-1 Promoter Slide modified from Alistair Forrest. Kanamori-Katayama, Itoh, Kawaji et al. 2011 Genome Research. “Unamplified cap analysis of gene expression on a single-molecule sequencer” Transcriptional Regulation A. TFBS prediction B. Co-expression TF A ×: Average expression No of CAGE tags In each promoter CAGE tags Gene B CAGE Promoter Gene C ● ● × ● ◆ ● ◆ × ■ ● × ■ ◆ ● × ■ ● × × ■ ■ ■ ◆ ◆ ◆ × ■ Gene D 0h TFBS prediction A: basis: TFBS prediction B: co-expression ◆ × 96h Co-expression = Total score TF A promoter B High High TF A promoter C High High TF A promoter D Low Low Motif Activity Response Analysis – MARA eps Genome Promoter1 m1 m1 m1 m2 m3 Promoter2 ・・・・ PromoterX m1 m1 m4 m5 Expression e ps m Rpm Ams Reaction efficiency • Number of possible binding sites Effective THP-1 cells are a monoblastic leukemia • Degree of conservation of cell the line motifwhich upon PMA treatment can differentiate into an concentration + + adherent• monocyte likestatus cell (CD14 , CSF1R ) Chromatin Suzuki, Forrest, van Nimwegen et al. Nature Genetics 2009, 41:5 Motif Activity Response Analysis • How much is a binding site used – Observed expression of all promoters over time – Predicted site count Suzuki, Forrest, van Nimwegen et al. Nature Genetics 2009, 41:5 NatGenet. Genet.2009 2009May;41(5):553-62. May;41(5):553-62. Nat Enhancers • Enhancers are sequence motifs • They bind factors (proteins) that are participating in the transcription initiation complex • Enhancers can be many kb away from the TSS • Insulators are acting in a similar way, but repressing expression • Is an enhancer a gene? Enhancer RNA • ENCODE reported (Nature, 489(7414), 101–108) – Enhancers identified by co-occurrence of H3K27ac and H3K4me1 ChIP-Seq data, centred on P300 binding sites, in HeLa cells • Enhancers make non-coding RNA Nature 465, 173–174 (2010). • Widespread transcription at neuronal activity-regulated enhancers. (Kim, T. K. et al. Widespread transcription at neuronal activityregulated enhancers. Nature 465, 182–187 (2010).) Djebali, S., Davis, C. A., Merkel, A., Dobin, A., Lassmann, T., Mortazavi, A., et al. (2012). Landscape of transcription in human cells. Nature, 489(7414), 101– 108. doi:10.1038/nature11233 RNA splicing in cancer http://en.wikipedia.org/wiki/RNA_splicing Example: Melanoma Transcriptome • discovery of aberrations that contribute to carcinogenesis • characterize the spectrum of cancerassociated mRNA alterations through integration of transcriptomic and structural genomic data – 11 novel melanoma gene fusions produced by underlying genomic rearrangements – 12 novel readthrough transcripts Genome Res. 2010 Apr;20(4):413-27 Melanoma Transcriptome: Gene Fusion Connecting genes located on different chromosomes! Melanoma Transcriptome: Gene Read-through • Genes fusions are ‘private’ – The same gene fusion was not observed in two melanoma patients (10 samples total) • Gene fusions in melanoma might not be the cancer causing events but consequences Chromosome Structure Ref: http://www.sequentiabiotech.com/ http://en.wikipedia.org/wiki/Chromosome_conformation_capture Mouse ES cells Dixon, J. R., Selvaraj, S., Yue, F., Kim, A., Li, Y., Shen, Y., et al. (2012). Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. doi:10.1038/nature11082 • Remote ER-a chromatin biding sites are anchored at gene promoters through long-range chromatin interactions • suggesting that ER-a functions by extensive chromatin looping to bring genes together for coordinated transcriptional regulation Nature. 2009 Nov 5;462(7269):58-64 Polymerase II Stalling stalled active No binding Nature Genetics 39, 1512 - 1516 (2007) • Pol II ChIP-chip in drosophila embryos • Stalled genes are highly enriched in developmental control genes Transcriptional Regulation in Cancer From observations to mechanisms • Observations => Biomarkers