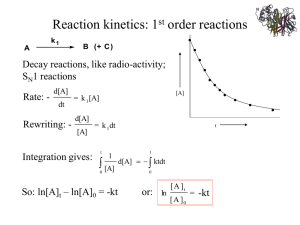
[S], K m
advertisement
![[S], K m](http://s2.studylib.net/store/data/005727372_1-436e65aa9ae986608afd0b1018baee47-768x994.png)
KAPITOLA 3 Enzymová katalýza I • katylytická aktivita enzymů • interakce enzym - substrát • koenzymy • vazebné místo pro substrát • fyzikálně chemické vlastnosti • nomenklatura enzymů Free energy, G Reaction coordinate The role of binding energy in catalysis. To lower the activation energy for a reaction, the system must acquire an amount of energy equivalent to the amount by which DG# is lowered. This energy comes largery from binding energy ( GB) contribued by formation of weak noncovalent interactions between substrate and enzyme in the transition state. The role of GB is analogous to that of GM. An imaginary enzyme (stickase) designed to catalyze the breaking of a metal stick. (a) To break, the stick must first be bent (the transition state). In the stickase, magnetic interactions take the place of weak-bonding interactions between enzyme and substrate. (b) An enzyme with a magnet-lined pocket complementary in structure to the stick (the substrate) will stabilize this substrate . Bending will be impeded by the magnetic attraction between stick and stickase. (c) An enzyme complementary to the reaction transition state will help to destabilize the stick, resulting in catalysis of the reaction. The magnetic interactions provide energy that compensates for the increase in free energy required to bend the stick. Reaction coordinate diagrams show the energetic consequences of complementarity to substrate versus complementarity to transition state. The term GM represents the energy contribued by the magnetic interactions between the stick and stickase. When the enzyme is complementary to the substrate, as in (b), the ES complex is more stable and has less free energy in the ground state than substrate alone. The result is an increase in the activation energy. For simplicity, the EP complexes are not shown. Many organic reactions are promoted by proton donors (general acids) or proton acceptors (general bases). The active sites of some enzymes contain amino acid functional groups, such as those shown here, that can participe in the catalytic process as proton donors or proton acceptors. Enzymes What? Proteins which catalyse biological reactions Properties? Highly specific Unchanged after reaction Can accelerate reactions >106 fold Turnover Cannot alter reaction equilibria Conversion of substrate to product: S P 2 factors determine the appearance of product: i. Thermodynamic ii. Kinetic Enzyme Classification Reaction profile for a simple reaction Rawn Biochemistry, International edition Reaction profile for an enzyme-catalysed reaction Uncatalysed Enzyme catalysed Rawn Biochemistry, International edition Enzyme kinetics Change in [S] and [P] with time 1200 Concentration (micromolar) 1000 800 S 600 400 P 200 0 0 50 100 150 200 Time (secs) 250 300 350 Impact of changing enzyme concentration Rate versus [enzyme] 3 Rate (v) micromolar per sec 2.5 2 1.5 1 0.5 0 0 1 2 3 4 [Enzyme] micromolar 5 6 Impact of changing substrate concentration Rate versus [substrate] 1 Rate (v) micomolar per sec 0.8 0.6 0.4 0.2 0 0 200 400 600 800 [Substrate] micromolar 1000 1200 Derivation of the Michaelis-Menten equation k1 k2 ES E+S E+P k -1 Rate of reaction: v k 2[ES] Equation (1) During the steady-state, [ES] is constant i.e. the rate of formation = rate of breakdown k 1[E][S] k 1[ES]k 2[ES] Therefore: k 1[E ][S ] [ES ] k 1 k2 k 1[E ][S ] [ES ] k 1 k2 If we introduce a constant KM defined as This equation becomes KM [E ][S] [ES ] KM k 1 k2 k1 Equation (2) The total concentration of enzyme introduced at the start of the reaction is still present, it is either bound to substrate or it is not, i.e. [E0] = [E] + [ES] Or rearranged: [E] = [E0] - [ES] Substituting this into Equation (2) gives us: ([E 0 ] [ES ])[S ] [ES ] KM [ES] = ([E0] - [ES])[S] KM Opening the bracket on the right-hand side gives: [ES] = [E0][S] - [ES][S] KM Therefore: KM [ES] = [E0][S] - [ES][S] Collect [ES]: KM [ES] + [S][ES] = [E0][S] (KM + [S])[ES] = [E0][S] Therefore: [ES] = [E0][S] [S] + KM [ES] = [E0][S] [S] + KM v k 2[ES] Substituting this back into equation (1): Gives us: v = k2 [E0][S] Equation (3) [S] + KM The maximum rate for the enzyme (Vmax) will be achieved when all of the Enzyme is saturated with substrate, i.e. [ES] = [E0] Putting this again into equation (1) we get: Vmax = k2 [E0] Notice that the term k2[E0] features in equation (3), which can now be written v= Vmax [S] KM + [S] Michaelis-Menten equation What does the KM actually tell us? Consider the case when the reaction rate (v) is half of the maximum rate (Vmax). Under these conditions the MM equation becomes: Vmax 2 = Vmax [S] KM + [S] This leaves us with: 2[S] = KM + [S] Which simplifies to: [S] = KM KM is the substrate concentration at which the reaction proceeds at half of the maximum rate (Vmax) Plot of rate (V) against substrate concentration ([S]) V max Initial rate, V V max/ 2 KM Substrate concentration, [S] v= Vmax [S] KM + [S] Taking the reciprocal of both sides can give 1 v = KM . Vmax This is in the general form 1 [S] 1 + Vmax y=mx+c i.e. a plot of 1/v against 1/[S] will give straight line This is a Lineweaver-Burk plot Lineweaver-Burk plot 1/V -1/K M slope = KM/ V max 1/V max 1/[S] KAPITOLA 4 Enzymová katalýza II • kinetika reakcí katalyzovaných enzymy • analýza reakční rychlosti • inhibice a regulace katalytické aktivity • metody studia Vmax 2 Vmax [ S ] Km [ S ] The Michaelis-Menten equation Dependence of initial velocity on substrate concentration, showing the kinetic parameters that define the limits of the curve at hight and low [S]. At low [S], Km>>[S], and the [S] term in the denominator of the Michaelis-Menten equation becomes insignificant; the equation simplifies to V0 = Vmax[S]/Km, and V0 exhibits a linear dependence on [S], as observed. At high [S], where [S]>>Km, the Km term in the denominator of the Michaelis-Menten equation becomes insignificant, and the equation simplifies to V0=Vmax; this is consistent with the plateau observed at high [S]. The Michaelis-Menten equation is therefore consistent with the observed dependence of V0 on [S], with the shape of the curve defined by the terms Vmax/Km at low [S] and Vmax at high [S]. 1 Km 1 1 V0 Vmax [ S ] Vmax The Lineweaver-Burk equation A double-reciprocal, or Lineweaver-Burk, plot. Common mechanisms for enzyme-catalyzed bisubstrate reactions. (a) the enzyme and both substrates come together to form a ternary complex. In ordered binding, substrate 1 must be bound before substrate 2 can bind productively. (b) an enzyme-substrate complex forms, a product leaves the complex, the altered enzyme forms a second complex with another substrate molecule, and the second product leaves, regenerating the enzyme. Substrate 1 may transfer a functional group to the enzyme (forming E´), which is subsequently transferred to substrate 2. This is a ping-pong or double-displacement mechanism. A B A shows a set of double-reciprocal plots obtained in the absence of the inhibitor and with two different concentrations of a competitive inhibitor. Increasing inhibitor concentration [I] results in the production of a family of lines with a common intercept on the 1/V0 axis but with different slopes. Because the intercept on the 1/V0 axis is equal to 1/Vmax, we can see that Vmax is unchanged by a presence of a competitive inhibitor. That is, regardless of the concentration of a competitive inhibitor, there is always some high substrate concentration that will displace the inhibitor from the enzyme´s active site. In noncompetitive inhibition, similar plots of the rate data give the family of lines shown in B, having a common intercept on the 1/[S] axis. This indicates that Km for the substrate is not altered by a noncompetitive inhibitor, but Vmax decreases. Three types of reversible inhibition. Competitive inhibitors bind to the enzyme´s active site. Noncompetitive inhibitors generally bind at a separate site. Uncompetitive inhibitors also bind at a separate site, but they bind only to the ES complex. KI is the equilibrum constant for inhibitor binding. Some well-studied examples of enzyme modification reactions. Allosteric Enzymes Obrázek Lehninger 3 obr grafy z PDF Lehninger str 228 Glutamine synthetase