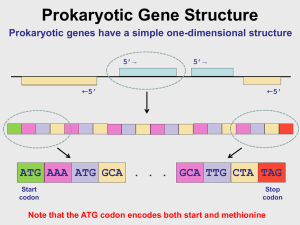
Finding substrings

Finding substrings
my $sequence = "gatgcaggctcgctagcggct";
#Does this string contain a startcodon?
if ($sequence =~ m/atg/) { print "Yes";
} else { print "No";
}
Finding substrings
my $sequence = "gatgcaggctcgctagcggct";
#Does this string contain a startcodon?
if ($sequence =~ m/atg/) { print "Yes";
} else { print "No";
}
=~ is a binding operator and means: perform the following action on this variable.
The following action m/atg/ in this case is a substring search, with the "m" for "match"' and substring "atg".
Finding substrings
my $sequence = "gatgcaggctcgctagcggct";
#Does this string contain a startcodon?
if ($sequence =~ m/atg/) { print "Yes";
} else { print "No";
}
If the substring occurs, the statement will return TRUE and the ifblock will be executed.
The value of $sequence does not change by the match.
Finding substrings, repeated
my $sequence = "gatgcaggctcgctagcggct"; my $count = 0; while($sequence =~ m/ggc/g) {
$count++;
} print "$count matches for gcc\n";
m//g
'g' option allows repeated matching, because the position of the last match is remembered
Finding substrings, repeated
my $sequence = "gatgcaggctcgctagcggct"; my $count = 0; while($sequence =~ m/ggc/g) {
$count++;
} print "$count matches for gcc\n";
Finding substrings, repeated
my $sequence = "gatgcaggctcgctagcggct"; my $codon = "ggc"; my $count = 0; while($sequence =~ m/$codon/g) {
$count++;
} print "$count matches for $codon\n";
Position after last match
my $sequence = "gatgcaggctcgctagcggct"; my $codon = "ggc"; print "looking for $codon from 0\n"; while($sequence =~ m/$codon/g) { print "found, will continue from: "; print pos($sequence),"\n";
}
Position after last match
my $sequence = "gatgcaggctcgctagcggct"; my $codon = "ggc"; pos($sequence) = 10; print "looking for $codon from 10\n"; while($sequence =~ m/$codon/g) { print "found, will continue from: "; print pos($sequence),"\n";
}
Replacing substrings
my $sequence = "gatgcagaattcgctagcggct"; print $sequence,"\n";
#Replace the EcoRI site with '******'
$sequence =~ s/gaattc/******/;
# gatgca******gctagcggct
#Replace all the other characters with space
$sequence =~ s/[^*]/ /g; print $sequence,"\n";
Output: gatgcagaattcgctagcggct
******
Examples of regular expressions
s/World/Wur/ replaces World with Wur, making "Hello World" "Hello Wur" s/t/u/ replaces the first 't' with 'u', "atgtag" becomes "augtag" s/t/u/g replaces all 't's with 'u's, "atgtag" becomes "auguag" s/[gatc]/N/g replaces all g,a,t,c's with N, "atgtag" becomes "NNNNNN" s/[^gatc]//g replaces all characters that are not g,a,t or c with nothing s/a{3}/NNN/g replaces all 'aaa' with 'NNN', "taaataa" becomes "tNNNtaa" m/sq/i match 'sq', 'Sq', 'sQ' and SQ: case insensitive m/^SQ/ match 'SQ' at the beginning of the string m/^[^S]/ match strings that do not begin with 'S' m/att?g/ match 'attg' and 'atg' m/a.g/ match 'atg', 'acg', 'aag', 'agg', 'a g', 'aHg' etc.
s/(\w+) (\w+)/$2 $1/ swap two words, "one two" => "two one" m/atg(…)*?(ta[ag]|tga)/ matches an ORF
The matched strings are stored
my $text = "This is a piece of text\n"; print $text;
$word = 0; while($text =~ /(\w+)\W/g) {
$word++; print "word $word: $1\n";
}
The matched strings are stored
my $text = "one two";
$text =~ /(\w+) (\w+)/g print "word one:$1 "; print "word two:$2 "; print "complete string: $&";
The matched strings are stored
my $sequence = "gatgcaggctcgctagcggct"; while ($sequence =~ m/([acgt]{3})/g) { print "$1\n";
}
Special characters
\t
\n tab newline
\r
\b return (CR)
"word" boundary not a "word" boundary \B
\w matches any single character classified as a "word" character
(alphanumeric or _)
\W matches any non-"word" character
\s matches any whitespace character (space, tab, newline)
\S matches any non-whitespace character
\d
\D matches any digit character, equiv. to [0-9] matches any non-digit character
\xhh character with hex. code hh
|
( )
[ ]
{ }
\
.
*
+
?
^
$
Metacharacters
beginning of string end of string any character except newline match 0 or more times match 1 or more times match 0 or 1 times; or shortest match alternative grouping, or storing set of characters repetition modifier quote or special
a* a+ a? a{m} a{m,} a{m,n} a{0,n}
Repetition
zero or more a's one or more a's zero or one a's (i.e., optional a) exactly m a's at least m a's at least m but at most n a's at most n a's
$mRNAsequence = "aaaauaaaaa";
$mRNAsequence =~ m/a{2,}ua{3,}/;
Greediness
Pattern matching in Perl by default is greedy, which means that it will try to match as much characters as possible. This can be prevented by appending the ? Operator to the expression
$sequence = "atgtagtagtagtagtag";
#This will replace the entire string: s/atg(tag)*//
#This will stop matching at the first tag: s/atg(tag)*?//
open SEQFILE, "example1.fasta"; my $sequence = ""; my $ID = <SEQFILE>; while (<SEQFILE>) { chomp;
$sequence .= $_;
} print $ID; print $sequence,"\n";
#SmaI striction (ccc^ggg)
$sequence =~ s/cccggg/ccc^ggg/g;
#PvuII striction (cag^ctg)
$sequence =~ s/cagctg/cag^ctg/g; my @sequenceFragments = split '\^', $sequence; print "\n", "-"x90, "\n"; print "Digested sequence:\n",$sequence,"\n\n"; print "-"x90,"\n"; print "Fragments:\n"; foreach $fragment(@sequenceFragments) { print $fragment,"\n"; print "-"x90,"\n";
}
>BTBSCRYR Bovine mRNA for lens beta-s-crystallin...
tgcaccaaacatgtctaaagctggaaccaaaattactttctttgaagacaaaaactttcaaggccgccactatgacagcgattgcgactgtgcagatttcc acatgtacctgagccgctgcaactccatcagagtggaaggaggcacctgggctgtgtatgaaaggcccaattttgctgggtacatgtacatcctaccccgg ggcgagtatcctgagtaccagcactggatgggcctcaacgaccgcctcagctcctgcagggctgttcacctgtctagtggaggccagtataagcttcagat ctttgagaaaggggattttaatggtcagatgcatgagaccacggaagactgcccttccatcatggagcagttccacatgcgggaggtccactcctgtaagg tgctggagggcgcctggatcttctatgagctgcccaactaccgaggcaggcagtacctgctggacaagaaggagtaccggaagcccgtcgactggggtgca gcttccccagctgtccagtctttccgccgcattgtggagtgatgatacagatgcggccaaacgctggctggccttgtcatccaaataagcattataaataa aacaattggcatgc
------------------------------------------------------------------------------------------
Digested sequence: tgcaccaaacatgtctaaagctggaaccaaaattactttctttgaagacaaaaactttcaaggccgccactatgacagcgattgcgactgtgcagatttcc acatgtacctgagccgctgcaactccatcagagtggaaggaggcacctgggctgtgtatgaaaggcccaattttgctgggtacatgtacatcctac ccc^g gg gcgagtatcctgagtaccagcactggatgggcctcaacgaccgcctcagctcctgcagggctgttcacctgtctagtggaggccagtataagcttcaga tctttgagaaaggggattttaatggtcagatgcatgagaccacggaagactgcccttccatcatggagcagttccacatgcgggaggtccactcctgtaag gtgctggagggcgcctggatcttctatgagctgcccaactaccgaggcaggcagtacctgctggacaagaaggagtaccggaagcccgtcgactggggtgc agcttccc cag^ctg tccagtctttccgccgcattgtggagtgatgatacagatgcggccaaacgctggctggccttgtcatccaaataagcattataaat aaaacaattggcatgc
------------------------------------------------------------------------------------------
Fragments: tgcaccaaacatgtctaaagctggaaccaaaattactttctttgaagacaaaaactttcaaggccgccactatgacagcgattgcgactgtgcagatttcc acatgtacctgagccgctgcaactccatcagagtggaaggaggcacctgggctgtgtatgaaaggcccaattttgctgggtacatgtacatcctacccc
-----------------------------------------------------------------------------------------ggggcgagtatcctgagtaccagcactggatgggcctcaacgaccgcctcagctcctgcagggctgttcacctgtctagtggaggccagtataagcttcag atctttgagaaaggggattttaatggtcagatgcatgagaccacggaagactgcccttccatcatggagcagttccacatgcgggaggtccactcctgtaa ggtgctggagggcgcctggatcttctatgagctgcccaactaccgaggcaggcagtacctgctggacaagaaggagtaccggaagcccgtcgactggggtg cagcttccccag
-----------------------------------------------------------------------------------------ctgtccagtctttccgccgcattgtggagtgatgatacagatgcggccaaacgctggctggccttgtcatccaaataagcattataaataaaacaattggc atgc
------------------------------------------------------------------------------------------
Exercises
6. Create a script to find the DNA fragments you get after cutting the sequence in the example1.fasta file with AluI and with AvaI
7. Find the open reading frames in the example1.fasta sequence
8. Translate the open reading frames to protein, using the standard genetic code from the
Geneticcode database
(http://srs.bioinformatics.nl)