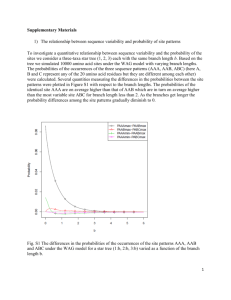
Termination in Translation
advertisement

Termination of Translation Chris Avins Elizabeth Durante Christine Noonan Basic Mechanism • 1. Elongation stops when a stop codon reaches A site of ribosome Basic Mechanism • 2. Release factor binds to stop codon Basic Mechanism • 3. Release factor adds H2O instead of an amino acid • 4. Bond is broken and polypeptide chain is freed tRNA Release Factors • Basic function: release polypeptide chain from mRNA • There are two release factors that work together in order to terminate translation (eRF1 & eRF3 in eukaryotes) (RF1/RF2 & RF3 in prokaryotes) Eukaryotic Release Factors • A stop codon is recognized by the heterodimer complex of eRF1 and eRF3 • These release factors mimic tRNA structurally and functionally – eRF3 triggers GTP hydrolysis, enhancing the rate of peptidyl release – It also recycles post-termination ribosomes to 5’ end to begin initiation again The Termination Signal • eRF1 recognizes all three stop codons • RF stop codon recognition is up to 60 times slower than sense codon decoding • eRF1 accurately discriminates between U-purinepurine codons and other sense codons, interacting with U-purine-purine codons stabilize eRF1 in a conformation to proceed to the next step The Termination Signal • Codons surrounding the stop codons are not random. The tetranucleotide is important in determining the efficiency of termination • As many as three succeeding nucleotides may contact the RF and play a role in termination • 5’ nucleotide context also influences efficiency Stop Codon Recognition • 8 protein residues found in eRF1 act in the physical interaction of eRF1 and mRNA stop codon that mediate stop codon recognition • 5 of the proteins were consistent in all analyzed species • 3 of the proteins were only the same in species using the same set of stop codons • 2 more proteins need more research • Stop codon selection in eukaryotes is not yet understood Prokaryotic Differences • The job done by eRF1 is divided between RF1 and RF2. Both discriminate between A&G at the 2nd and 3rd positions of stop codons using the PAT & SPF tripeptides • Peptidyl release is independent of RF3 • RF3 stimulates the termination rxn. and binds guanine nucleotides but is not codon-specific • There is no structural resemblance between RF1/2 and RF3 and eRF1 and eRF3 due to evolutionary origin of translation termination References • http://www.ncbi.nlm.nih.gov/pmc/articles/PM C394485/pdf/emboj00040-0223.pdf • http://mic.sgmjournals.org/cgi/reprint/147/2/25 5 • http://www.nature.com/emboj/journal/v22/n7/f ull/7595057a.html • http://www.springerlink.com/content/lx223777 867w3521/fulltext.pdf