Dave Vanness, PhD An Introduction to Bayesian GLM Methods Research Scientist
advertisement
An Introduction to Bayesian GLM Methods
for CEA with Individual Data
Dave Vanness, PhD
Research Scientist
Center for Health Economics & Science Policy
Overview/Learning Objectives
Introduction to Bayesian Analysis
Bayes’ Rule
Markov Chain Monte Carlo
Analytical Example
Hypothetical CEA alongside a trial (simulated dataset based on MEPS
data)
Bayesian Generalized Linear Models (GLM) of:
Potential latent class of individuals with different response to treatment
Cure and Adverse Events - Bernoulli with Probit Link
Cost - Gamma with Log Link
QALY - Beta with Logistic Link
Counter-factual Simulation Analysis of Results
Goal: The probability that a treatment is cost-effective
Posterior ICER, expected net benefit and acceptability
Basics of Bayesian Analysis
Basics of Bayesian Analysis
Y : Data –
events we
observe
θ : Parameter –
probability of
event
Bayes’ Rule
Likelihood
Posterior
P(θ | Y ) =
Prior
L(Y | θ ) × P (θ )
θ
θ
θ
L
Y
P
d
(
|
)
×
(
)
∫
Normalizing Constant
is proportional to …
P(θ | Y ) α L(Y | θ ) × P(θ )
What you know now
times what you knew before
(PRIOR)
(POSTERIOR)
the likelihood of what
you just observed
A simple binomial example
Consider a treatment that can either succeed (Yi=1) or
fail (Yi=0) for individual i.
Let θ be the unknown average success rate for that
treatment.
No Prior Knowledge
Suppose you know absolutely nothing about the
treatment success rate before observing a new set of
data (yet to be collected).
What might your (lack of) prior knowledge look like if
you plotted it out?
Suppose instead that you had a lot of
knowledge about the treatment success
rate θ and were almost certain treatment
is between 60% and 70% successful.
What might that look like?
How do we get from this …
to this?
With data and a model…
Suppose we observe one patient who has undergone
treatment.
Suppose that patient failed (rats!):
Y1 = 0
The Likelihood Function (model)
L(Y1 = 0 | θ)
1
0
0
1
θ
Applying Bayes’ Rule
P(θ)
L(Y1 = 0 |θ)
1
1
X
0
0
1
θ
0
0
1
θ
The Posterior
P(θ|Y1=0)
2
0
0
1
θ
If at first you don’t succeed …
This time, we observe patient 2, whose treatment is a
success (yay!)
Y2 = 1
Now, we bring information with us.
Our posterior P(θ|Y1=0) becomes our new
prior.
P(θ)
L(Y2 = 1 |θ)
1
2
X
0
0
1
Our non-flat prior…
θ
θ
0
0
1
The likelihood of
observing a success.
P(θ|Y2=1,Y1=0)
1
0
0
1
θ
The Bayesian Learning Process
Heterogeneity
This was a very simple (homogenous) model: every
individual’s outcome is drawn from the same
distribution.
The extreme in the opposite direction (complete
heterogeneity) is also fairly simple:
Yi ~ Bernoulli(θi)
But it’s pretty difficult to extrapolate (make predictions)
when there is no systematic variation.
Modeling Heterogeneity
Usually, we assume there is a relationship that explains
some heterogeneity in observed outcomes.
The classical normal regression model:
Yi = Xiβ + εi
Xi is a row vector of individual covariates
β is a column vector of parameters
εi ~ N(0,σ2)
We can also write this as:
Yi ~ N(µi,,σ2), µi = Xiβ [likelihood]
θ = (β, σ2) ~ P(θ) [prior]
The posterior we are interested in is the product of the likelihood
and prior.
We will not be able to calculate this analytically
Markov Chain Monte Carlo (MCMC)
Good text is Markov Chain Monte Carlo in Practice by
Gilks, Richardson and Spiegelhalter (Chapman & Hall)
Goal is to construct an algorithm that generates random
draws from a distribution that converges to P(θ|X).
We then collect those draws and analyze them (take
their mean, median, etc., or run them through as
parameters of a cost-effectiveness simulation, etc.).
Gibbs Sampling
When θ is multidimensional, it can be useful to break
down the joint distribution P(θ|X) into a sequence of “full
conditional distributions”
P(θj|θ-j,X) = L(X|θj,θ-j) P(θj|θ-j)
where “-j” signifies all elements of θ other than j.
We can then specify a starting vector θ-j0 and, if P(θj|θj,X) is not from a known type of distribution, we can use
the Metropolis algorithm to sample from it.
Running from j = 1 to M gives one full sample of θ.
θ2
θ1
θ2
θ02
θ1
θ2
θ02
θ1
θ2
θ02
0
θ01
θ1
θ2
θ02
0
θ01
θ1
θ2
θ02
0
θ12
θ01
θ1
θ2
θ02
0
θ12
θ01
θ1
θ2
θ02
θ12
0
1
θ11
θ01
θ1
θ2
θ02
0
4
3
1
2
θ1
http://www.mrc-bsu.cam.ac.uk/bugs/welcome.shtml
Using MCMC to Conduct CEA
Using the flexibility of WinBUGS, we can explore the
relationships among treatments, covariates, costs and
health outcomes using Generalized Linear Models
(GLM) and perform probabilistic sensitivity analysis at
the same time.
Obtain Markov Chain Monte Carlo samples from the posterior
distribution of unknown GLM model parameters, given our data,
model and prior beliefs.
These "draws" can be used not simply to conduct inference (the
"credible interval") but also to perform uncertainty analysis.
For an introduction to the general approach, see O'Hagan A and
Stevens JW. A framework for cost-effectiveness analysis from
clinical trial data. Health Economics, 2001; 10(4): 303-315.
Simulated CEA Dataset
800 individuals selected at random from 2,452
individuals who self-reported hypertension in the 2005
MEPS-HC
Covariates were:
Age
Sex (Male = 1)
BMI
We created a latent class that equals 1 if an individual
self-reported diabetes; 0 otherwise. The class variable
was excluded from all analysis (assumed to be
unobservable)
Simulated CEA Dataset
Treatment (T = 1) assigned randomly (no treatment
selection bias)
Ti ~ Bernoulli (0.5)
Adverse events (AE = 1) : can occur in anyone, but will
occur with probability 1 for individuals in the latent class
who receive treatment.
AEi ~ Bernoulli (PiAE)
PiAE = 0.1 + 0.9*Ti*Classi
“Cure” (S = 1) : 8 times more likely with treatment than
without and does not depend on class.
Si ~ Bernoulli (PiS)
PiS = 0.8*Ti + 0.1*(1-Ti)
Simulated CEA Dataset
Costs are increasing in age and are higher for the
treated and people with AEs, lower if treatment is
successful:
Ci = CiT*Ti + CiX
CiT ~ Gamma(25,1/400)
CiX ~ Gamma(4,1/exp(XiβC))
where Xi is a row vector consisting of: 1~Agei~Sexi~AEi~Si and
βC is a column vector of parameters: 7|.03|0|1.5|-.5.
Simulated Costs
Simulated CEA Dataset
QALYs are decreasing in age and are lower for males
and people with AEs and higher for people with
successful treatment:
Qi ~ Beta(αi,βi)
αi = βi exp(XiβQ)
βi = 1.2 + 2*Ti
where Xi is a row vector consisting of: 1~Agei~Sexi~AEi~Si and
βQ is a column vector of parameters:
1|-.01|.25|-1|1.5
Simulated QALYs
A simple CEA
Sample (n=800) ICER: $74,357/QALY
Population (n=2452) ICER: $79,933/QALY
Population ICER by (unobserved class):
Class 0 (no diabetes): $43,519/QALY
Class 1 (diabetes): $589,954/QALY
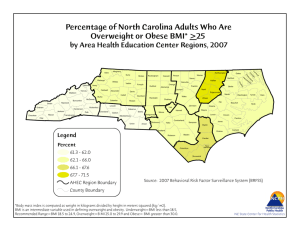
Latent Class and BMI
Estimating Bayesian GLM Models
S ~ Bernoulli(Pis)
Pis = Φ(Xisβs)
Xis = 1~Agei~BMIi~Sexi~Ti~Ti*(Agei~BMIi~Sexi)
AE ~ Bernoulli(PiAE)
PiAE = Φ(XiAEβAE)
XiAE = 1~Agei~BMIi~Sexi~Ti~Ti*(Agei~BMIi~Sexi)
Note: we are using non-informative (“flat”) priors, which give results
comparable to Maximum Likelihood. But we could bring outside
information into the prior (from other related trial or observational
data, meta analysis, expert opinion, etc).
BUGS Code (Probits)
#Treatment Success and Adverse Event Model
for ( i in 1 : 800 ){
S[i] ~ dbern(p_S[i])
p_S[i] <- phi(arg.S[i])
arg.S[i] <- max(min(S_CONSTANT + S_AGE*st_AGE[i] +
S_MALE*MALE[i] + S_BMI*st_BMI[i] + S_T*T[i]
+ S_TxAGE*T[i]*st_AGE[i] +
S_TxMALE*T[i]*MALE[i]
+ S_TxBMI*T[i]*st_BMI[i],5),-5)
AE[i] ~ dbern(p_AE[i])
p_AE[i] <- phi(arg.AE[i])
arg.AE[i] <- max(min(AE_CONSTANT + AE_AGE*st_AGE[i]
+ AE_MALE*MALE[i] + AE_BMI*st_BMI[i] +
AE_T*T[i] + AE_TxAGE*T[i]*st_AGE[i] +
AE_TxMALE*T[i]*MALE[i] +
AE_TxBMI*T[i]*st_BMI[i],5),-5)
}
BUGS Code (Probits)
# Priors
# Success coefficients
S_CONSTANT ~ dnorm(0,.0001)
S_AGE ~ dnorm(0,.0001)
S_MALE ~ dnorm(0,.0001)
S_BMI ~ dnorm(0,.0001)
S_T ~ dnorm(0,.0001)
S_TxAGE ~ dnorm(0,.0001)
S_TxMALE ~ dnorm(0,.0001)
S_TxBMI ~ dnorm(0,.0001)
# Adverse event coefficients
AE_CONSTANT ~ dnorm(0,.0001)
AE_AGE ~ dnorm(0,.0001)
AE_MALE ~ dnorm(0,.0001)
AE_BMI ~ dnorm(0,.0001)
AE_T ~ dnorm(0,.0001)
AE_TxAGE ~ dnorm(0,.0001)
AE_TxMALE ~ dnorm(0,.0001)
AE_TxBMI ~ dnorm(0,.0001)
Draws from the MCMC sampler for treatment Adverse
Event Parameter
Draws from the MCMC sampler for Treatment x BMI
Adverse Event parameter
Another way of looking at the draws: Posterior
Density
Posterior Credible Interval
Node
Adverse Events
(Bernoulli - Probit)
AGE
BMI
CONSTANT
MALE
T
TxAGE
TxBMI
TxMALE
Success (Bernoulli Probit)
AGE
BMI
CONSTANT
MALE
T
TxAGE
TxBMI
TxMALE
Posterior
Mean
Posterior
Standard
Deviatio
n
Monte
Carlo
Error
2.50%
median
97.50%
0.0094
-0.0119
-1.1630
-0.0121
0.6391
0.0608
0.2338
-0.0047
0.0783
0.0783
0.1181
0.1690
0.1487
0.1023
0.1043
0.2211
0.0025
0.0029
0.0054
0.0080
0.0069
0.0034
0.0037
0.0104
-0.1363
-0.1665
-1.4030
-0.3376
0.3501
-0.1457
0.0342
-0.4334
0.0090
-0.0095
-1.1610
-0.0135
0.6405
0.0620
0.2353
-0.0014
0.1647
0.1339
-0.9381
0.3178
0.9348
0.2571
0.4395
0.4152
-0.0022
-0.0012
-1.3500
0.1391
2.2460
0.0326
-0.1557
-0.2274
0.0885
0.0859
0.1214
0.1687
0.1519
0.1156
0.1131
0.2189
0.0031
0.0031
0.0055
0.0077
0.0068
0.0039
0.0040
0.0097
-0.1753
-0.1764
-1.5990
-0.1796
1.9600
-0.1936
-0.3741
-0.6529
-0.0013
0.0002
-1.3490
0.1384
2.2480
0.0336
-0.1560
-0.2242
0.1693
0.1633
-1.1180
0.4846
2.5600
0.2552
0.0684
0.2165
GLM Cost Model
We model cost as a mixture of Gammas (separate distributions of
cost with and without treatment).
Ci = Ti*Ci1 + (1-Ti)*Ci0
Ci1 ~ Gamma(shapeC1,scaleiC1)
Ci0 ~ Gamma(shapeC0,scaleiC0)
Using log function to link mean cost to shape and scale
parameters.
Key: mean of a gamma distribution = shape/scale…
Ln[mean(Ci)] = Xiβ
exp(Ln[mean(Ci)]) = exp(Xiβ)
mean(Ci) = exp(Xiβ)
shape/scalei = exp(Xiβ)
scalei = shape/exp(Xiβ)
Posterior Credible Interval
Posterior
Mean
Posterior
Standard
Dev.
Monte
Carlo
Error
2.50%
median
97.50%
AE
1.5190
0.0737
0.0015
1.3770
1.5190
1.6690
AGE
0.1552
0.0240
0.0004
0.1098
0.1550
0.2037
BMI
0.0031
0.0230
0.0004
-0.0424
0.0032
0.0477
CONSTANT
-0.8600
0.0358
0.0012
-0.9289
-0.8600
-0.7893
MALE
0.0277
0.0490
0.0015
-0.0671
0.0289
0.1231
S
-0.5440
0.0775
0.0016
-0.6897
-0.5430
-0.3867
SHAPE
4.5590
0.3130
0.0029
3.9700
4.5530
5.1890
AE
0.7035
0.0253
0.0006
0.6544
0.7035
0.7544
AGE
0.0746
0.0114
0.0002
0.0522
0.0747
0.0971
BMI
0.0147
0.0121
0.0002
-0.0080
0.0144
0.0390
CONSTANT
0.2002
0.0303
0.0015
0.1382
0.2013
0.2563
MALE
0.0016
0.0229
0.0006
-0.0420
0.0015
0.0472
S
-0.1725
0.0299
0.0014
-0.2294
-0.1736
-0.1133
SHAPE
18.3500
1.2890
0.0119
15.9000
18.3300
20.9600
Node
Cost w/o Treatment (GammaLog)
Cost w/Treatment (Gamma Log)
GLM QALY Model
Rescale Q to [0,1] interval by dividing by maximum
possible Q (follow-up time)
Qi = Ti*Qi1 + (1-Ti)*Qi0
Qi1 ~ Beta(aiQ1, bQ1)
Qi0 ~ Beta(aiQ0, bQ0)
We use the logit function to link mean QALYs to shape
and scale parameters.
Key: mean of a Beta distribution = a/(a+b)
mean(Qi) = exp(Xiβ)/(1+exp(Xiβ))
mean(Qi) = ai/(ai + b)
ai + b exp(Xβ) = ai exp(Xiβ) + b exp(Xiβ)
ai = b exp(Xiβ)
Posterior Credible Interval
Node
Posterior
Posterior Standard
Mean
Dev.
Monte
Carlo
Error
2.50%
median
97.50%
QALY w/o Treatment (Beta
- Logit)
AE
AGE
BMI
CONSTANT
MALE
S
β
-1.1760
-0.0193
-0.0015
0.5294
0.1366
1.3100
1.1600
0.1578
0.0493
0.0479
0.0733
0.0963
0.1615
0.0756
0.0030
0.0009
0.0010
0.0023
0.0030
0.0030
0.0008
-1.4950
-0.1144
-0.0954
0.3849
-0.0565
0.9928
1.0170
-1.1740
-0.0205
-0.0017
0.5300
0.1368
1.3160
1.1580
-0.8751
0.0795
0.0927
0.6753
0.3198
1.6240
1.3110
QALY w/Treatment (Beta Logit)
AE
AGE
BMI
CONSTANT
MALE
S
β
-0.9542
-0.0092
0.0865
0.4160
0.2812
1.4720
3.4980
0.0677
0.0300
0.0305
0.0797
0.0577
0.0799
0.2322
0.0015
0.0006
0.0006
0.0041
0.0017
0.0040
0.0022
-1.0880
-0.0673
0.0298
0.2625
0.1621
1.3120
3.0580
-0.9524
-0.0088
0.0862
0.4129
0.2827
1.4760
3.4950
-0.8262
0.0484
0.1478
0.5812
0.3906
1.6230
3.9740
Counterfactual Simulations
Within WinBUGS, we take the draws from the
parameter posteriors and, for a hypothetical individual
(X profile):
Assign to No Treatment (T = 0)
Assign to Treatment (T = 1)
Simulate cure or no cure
Simulate adverse event
Simulate cost and QALY, given simulated cure and adverse event
status
Repeat cure, adverse event, cost and QALY simulations
Repeat, say, 1,000 times and calculate average incremental
costs and QALYs.
The following ICERs apply to a female (Sexi = 0) of
average age (z-transformed Agei = 0) at 5 different
levels of z-transformed BMIi = {-2,-1,0,1,2}
Posterior ICERs
$50K
BMI z-score
-2.0
-1.0
0.0
1.0
2.0
Posterior Acceptability (BMI = -2)
(WTP from $0 to $200,000)
Posterior Acceptability (BMI = 2)
(WTP from $0 to $200,000)
Posterior Expected Net Benefit
(WTP from $0 to $200,000)
BMI z-score
-2.0
BMI = -2
-1.0
0.0
1.0
2.0
BMI = -1
BMI = 0
BMI = 1
BMI = 2
WTP Thresholds
$50K
$60K
$70K
$90K
$120K
Conclusion
Advantages to Bayesian Approach
Statistical analysis is coherent with rational decision-making under
uncertainty
Inference is on parameters, not future data
Therefore, can hypothesize about arbitrary “value functions” of parameters –
like cost-effectiveness or net benefit
Posterior draws are ready-made for inclusion in decision-analytic models
MCMC is a great tool for estimating complicated models
Probabilistic sensitivity analysis parameters will have correct interparameter
correlation if model is correctly specified
Very flexible and robust in WinBUGS
Many convergence diagnostics, including nice visualization
Can incorporate “prior information” from previous related studies, metaanalysis, etc. Comes under the general topic of “evidence-synthesis.”
Conclusion
Drawbacks to Bayesian approach
Still controversial: resistance among some journals and
reviewers.
However, Bayesian methods are playing a key role in submissions
to NICE (e.g., mixed treatment comparison meta-analysis)
Choice of prior is not always clear
Informative or “non-informative” may matter if observed data are
sparse
Some models are sensitive to what seem to be trivial differences in
non-informative priors (e.g., rare events models)
WinBUGS has a steep learning curve – “classical” MLE methods
are easier to implement, say, in STATA
References
George Woodworth’s book “Biostatistics: A Bayesian Introduction” (Wiley-Interscience, 2004 ISBN: 0471468428
9780471468424 has an excellent WinBUGS tutorial (Appendix B), the text of which may be found here:
http://www.stat.uiowa.edu/~gwoodwor/BBIText/AppendixBWinbugs.pdf
Carlin BP, TA Louis. Bayes and Empirical Bayes Methods for Data Analysis. 2nd Edition, London: Chapman & Hall,
2000.
Claxton K. The irrelevance of inference: a decision-making approach to the stochastic evaluation of health care
technologies. J.Health Econ. 1999 Jun;18(3):341-364.
Fryback DG, NK Stout, MA Rosenberg. An Elementary Introduction to Bayesian Computing Using WinBUGS.
International Journal of Technology Assessment in Health Care, 2001;17(1):98-113.
Gelman A and J Hill. Data Analysis Using Regression and Multilevel/Hierarchical Models. Cambridge: Cambridge
University Press, 2007.
Gilks WR, S Richardson, DG Spiegelhalter. Markov Chain Monte Carlo in Practice. London: Chapman & Hall, 1996.
Hastie T, R Tibshirani and J Friedman. The Elements of Statistical Learning. Data Mining, Inference and Prediction.
Springer, 2002.
Luce BR, Claxton K. Redefining the analytical approach to pharmacoeconomics. Health Econ. 1999 May;8(3):187-189.
Lunn DJ, Thomas A, Best N, Spiegelhalter D. WinBUGS-A Bayesian modelling framework: Concepts, structure, and
extensibility. Statistics and Computing 2000;10(4):325-337.
O'Hagan A, Stevens JW, Montmartin J. Bayesian cost-effectiveness analysis from clinical trial data. Stat.Med. 2001
Mar 15;20(5):733-753.
Skrepnek GH. The contrast and convergence of Bayesian and frequentist statistical approaches in pharmacoeconomic
analysis. Pharmacoeconomics 2007;25(8):649-664.
Spiegelhalter DJ, Myles JP, Jones DR, Abrams KR. Bayesian methods in health technology assessment: a review.
Health Technol.Assess. 2000;4(38):1-130.
Tanner MA. Tools for Statistical Inference. Methods for the Exploration of Posterior Distributions and Likelihood
Functions. 3d ed. Springer, 1996.
Vanness DJ and Kim WR. “Empirical Modeling, Simulation and Uncertainty Analysis Using Markov Chain Monte Carlo:
Ganciclovir Prophylaxis in Liver Transplantation,” Health Economics, 2002: 11(6), 551-566.