This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike License. Your use of this
material constitutes acceptance of that license and the conditions of use of materials on this site.
Copyright 2006, The Johns Hopkins University and Karl W. Broman. All rights reserved. Use of these materials
permitted only in accordance with license rights granted. Materials provided “AS IS”; no representations or
warranties provided. User assumes all responsibility for use, and all liability related thereto, and must independently
review all materials for accuracy and efficacy. May contain materials owned by others. User is responsible for
obtaining permissions for use from third parties as needed.
ANOVA assumptions
• Data in each group are a random sample from
some population.
• Observations within groups are independent.
• Samples are independent.
• Underlying populations normally distributed.
• Underlying populations have the same variance.
Diagnostics
• QQ plot within each group
• QQ plot of all residuals, yti − ȳt·
• Plot residuals, yti − ȳt·, against fitted values, ȳt·.
• Plot SD versus mean for each group.
• Plot the residuals against other factors.
(e.g., order of measurements, weight or age of mouse).
A
B6
1
2
4
5
6
7
8
10
11
12
13
14
15
17
18
19
24
25
26
Strain
Strain
Example
0
1000
2000
3000
4000
5000
A
B6
1
2
4
5
6
7
8
10
11
12
13
14
15
17
18
19
24
25
26
6000
2.5
IL10 response
3.0
3.5
log10 IL10 response
ANOVA Tables
Original scale / 1000:
source
SS
df
MS
between strains
33
20 1.69 1.70
within strains
124 125 0.99
total
157 145
log10 scale:
F P-value
0.042
source
SS
df
MS
between strains
3.35
within strains
9.29 125 0.074
total
F
P
20 0.167 2.25 0.0036
12.63 145
A
B6
1
2
4
5
6
7
8
10
11
12
13
14
15
17
18
19
24
25
26
Strain
Strain
Residuals
−1000
0
1000
2000
3000
residuals (IL10)
4000
5000
A
B6
1
2
4
5
6
7
8
10
11
12
13
14
15
17
18
19
24
25
26
−0.5
0.0
residuals (log10 IL10)
0.5
Within-group QQ-plots : IL10
Strain A
Strain B6
Strain 2
2500
1800
1500
1000
1600
2000
Sample Quantiles
Sample Quantiles
Sample Quantiles
2000
1500
1000
1400
1200
1000
800
600
500
−1.5
−1.0
−0.5
0.0
0.5
1.0
1.5
−1.5
−1.0
Theoretical Quantiles
Strain 4
−0.5
0.0
0.5
1.0
1.5
−1.5
−1.0
−0.5
0.0
0.5
Theoretical Quantiles
Theoretical Quantiles
Strain 8
Strain 12
1.0
1.5
1.0
1.5
1.0
1.5
1.0
1.5
3500
5000
3000
4000
3000
2000
Sample Quantiles
Sample Quantiles
Sample Quantiles
3000
2500
2000
1500
−1.0
−0.5
0.0
0.5
1.0
1.5
1500
500
500
−1.5
2000
1000
1000
1000
2500
−1.5
−1.0
Theoretical Quantiles
−0.5
0.0
0.5
1.0
1.5
−1.5
−1.0
Theoretical Quantiles
−0.5
0.0
0.5
Theoretical Quantiles
Within-group QQ-plots : log10 IL10
Strain A
Strain B6
3.4
Strain 2
3.4
3.2
3.0
2.8
Sample Quantiles
Sample Quantiles
Sample Quantiles
3.3
3.2
3.2
3.1
3.0
2.9
3.1
3.0
2.9
2.8
2.8
2.7
−1.5
−1.0
−0.5
0.0
0.5
1.0
1.5
−1.5
−0.5
0.0
0.5
1.0
1.5
−1.5
−1.0
−0.5
0.0
0.5
Theoretical Quantiles
Theoretical Quantiles
Theoretical Quantiles
Strain 4
Strain 8
Strain 12
3.6
3.4
3.4
3.2
Sample Quantiles
3.4
Sample Quantiles
Sample Quantiles
−1.0
3.2
3.0
3.2
3.0
2.8
2.8
3.0
−1.5
−1.0
−0.5
0.0
0.5
Theoretical Quantiles
1.0
1.5
−1.5
−1.0
−0.5
0.0
0.5
Theoretical Quantiles
1.0
1.5
−1.5
−1.0
−0.5
0.0
0.5
Theoretical Quantiles
QQ plots of all residuals
log10 IL10
IL10
−1000
0
1000
2000
3000
4000
5000
−0.5
0.0
Residuals
0.5
Residuals
4000
Sample Quantiles
Sample Quantiles
5000
3000
2000
1000
0
0.5
0.0
−0.5
−1000
−2
−1
0
1
2
−2
−1
Theoretical Quantiles
0
1
2
Theoretical Quantiles
Residuals vs fitted values
5000
4000
0.5
residuals (log10 IL10)
residuals (IL10)
3000
2000
1000
0
0.0
−0.5
−1000
500
1000
1500
fitted values (IL10)
2000
2.7
2.8
2.9
3.0
3.1
fitted values (log10 IL10)
3.2
3.3
SDs vs means
0.5
0.4
SD (log10 IL10)
SD (IL10)
1500
1000
0.3
0.2
500
0.1
500
1000
1500
2000
2.7
2.8
Mean (IL10)
2.9
3.0
3.1
3.2
3.3
Mean (log10 IL10)
Homogeneity of variances
One of the ANOVA assumptions was homogeneity of the group
variances. This can formally be tested with Bartlett’s test.
Assume we have k treatment groups.
nt
number of cases in treatment group t.
N
number of cases (overall).
Yti
response i in treatment group t.
Ȳt·
average response in treatment group t.
S2t
the sample variance in treatment group t.
Bartlett’s test
We want to test
H0 : σ12 = · · · = σk2
versus
Ha : H0 is false.
• Calculate the pooled sample variance:
P
P
(nt – 1) × S2t
(nt – 1) × S2t
2
tP
= t
S =
N–k
t (nt – 1)
• Calculate the test statistic
X 2 = (N – k) × log(S2 ) –
X
(nt – 1) × log(S2t)
t
• Calculate the following correction factor:
#
"
X 1
1
1
–P
C=1+
3(k – 1)
nt – 1
t (nt – 1)
t
If H0 is true, then
X 2/C ∼ χ2(df=k–1)
Example
• For the example data, there are 21 strains with between 5 and 10 observations
per strain.
• The pooled sample variance on original scale / 1000 is 0.99.
• The pooled sample variance on log10 scale is 0.074.
• The test statistics were 79.9 and 34.0.
• The correction factor ended up being 1.07.
• Thus we look at the values 79.9 / 1.07 = 74.8 and 34.0 / 1.07 = 31.8.
• Since there are 21 strains, we refer to the χ2(df = 20) distribution.
• We end up with P-values of 2.9 × 10–8 and 0.045.
The R function bartlett.test() can be used to do these calculations.
Hartley’s F-max test
In case that the number of observations are the same in every
treatment group, there is a quick and dirty alternative to Bartlett’s
test, called Hartley’s F-max test. For this test, simply compute
Fmax =
max(S2t)
min(S2t)
There is a look-up table with critical values for Fmax, using the number of treatment groups (k) and the degrees of freedom associated
with each of the group variances (nt – 1).
Number of treatment groups
Df
α
2
3
2
0.05
39
0.01
199
0.05
15.4 27.8 39.2 50.7
62
72.9
83.5
93.9
104
114
124
0.01
47.5 85
151
184
216
249
281
310
337
361
0.05
9.6
15.5 20.6 25.2
29.5
33.6
37.5
41.4
44.6
48
51.4
0.01
23.2 37
69
79
89
97
106
113
120
0.05
7.15 10.8 13.7 16.3
18.7
20.8
22.9
24.7
26.5
28.2
29.9
0.01
14.9 22
38
42
46
50
54
57
60
0.05
5.82 8.38 10.4 12.1
13.7
15
16.3
17.5
18.6
19.7
20.7
0.01
11.1 15.5 19.1 22
25
27
30
32
34
36
37
0.05
4.99 6.94 8.44 9.7
10.8
11.8
12.7
13.5
14.3
15.1
15.8
0.01
8.89 12.1 14.5 16.5
18.4
20
22
23
24
26
27
0.05
4.43 6
7.18 8.12
9.03
9.8
10.5
11.1
11.7
12.2
12.7
0.01
7.5
11.7 13.2
14.5
15.8
16.9
17.9
18.9
19.8
21
0.05
4.03 5.34 6.31 7.11
7.8
8.41
8.95
9.45
9.91
10.3
10.7
0.01
6.54 8.5
11.1
12.1
13.1
13.9
14.7
15.3
16
16.6
0.05
3.72 4.85 5.67 6.34
6.92
7.42
7.87
8.28
8.66
9.01
9.34
0.01
5.85 7.4
10.4
11.1
11.8
12.4
12.9
13.4
13.9
3
4
5
6
7
8
9
10
5
6
7
8
9
10
11
12
87.5 142
202
266
333
403
475
550
626
704
448
1036 1362 1705 2063 2432 2813 3204 3605
9.9
4
729
120
49
28
9.9
8.6
59
33
9.6
Another example
Rate of growth in fish eggs from different mothers
360
tth
340
320
300
280
1
2
3
4
5
6
7
8
mom
8
7
6
mom
5
4
3
2
1
280
300
320
340
tth
360
ANOVA Table
source
SS
df
between moms
12757
within moms
73510 546
total
86267 553
MS
F P-value
7 1822 13.5
4e-16
135
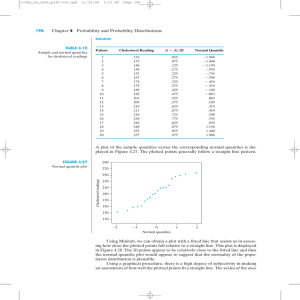
QQ plot of all residuals
40
Residuals
20
0
−20
−40
−3
−2
−1
0
Normal quantiles
1
2
3
QQ plots within each group
Mom 2
340
350
330
340
330
350
340
340
330
320
310
300
320
Mom 4
Residuals
360
Mom 3
Residuals
370
350
Residuals
Residuals
Mom 1
320
−2
−1
0
1
2
−2
−1
0
1
290
2
−2
−1
0
1
2
−2
−1
0
1
Normal quantiles
Normal quantiles
Normal quantiles
Normal quantiles
Mom 5
Mom 6
Mom 7
Mom 8
350
330
330
320
2
360
335
330
300
325
320
290
Residuals
310
Residuals
340
Residuals
Residuals
310
300
300
280
300
320
310
290
310
330
320
310
330
320
310
300
315
300
290
−2
−1
0
1
Normal quantiles
2
−1.5
−0.5
0.5
Normal quantiles
1.0
1.5
−2
−1
0
1
Normal quantiles
2
−2
−1
0
1
Normal quantiles
2