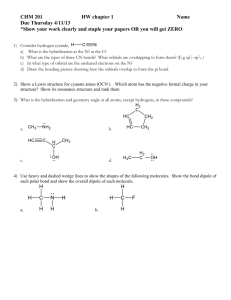
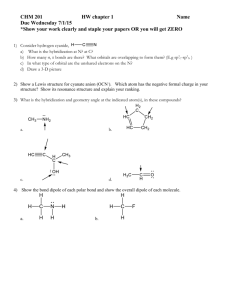
Hybridization
advertisement

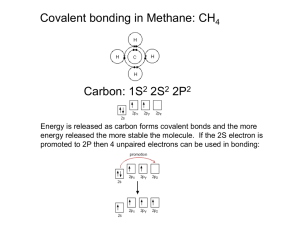
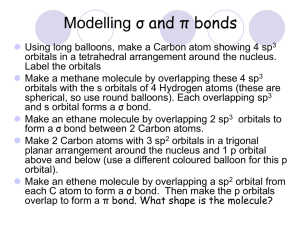
COVALENT BONDING: ORBITALS Chapter 9 Hybridization The mixing of atomic orbitals to form special molecular orbitals for bonding. The atoms are responding as needed to give the minimum energy for the molecule. Molecular Geometry & Hybridization Parent geometry determines the hybridization. Molecular structure is the actual geometry. Energy-level diagram showing the formation of four sp3 hybrid orbitals. 09_158 z z x x y y s z py x pz x y z x y z Hybridization y px z x y z z x y x y sp3 gives a tetrahedral arrangement sp3 sp3 sp3 One 2s and three 2p orbitals hybridize to form a new set of sp3 hybrid orbitals. sp3 hybrid orbital • 4 effective electron pairs. • tetrahedral geometry. • 109.5 o bond angle. 09_161 H1s sp3 sp3 H1s C H1s sp3 sp3 H1s The tetrahedral set of four sp3 orbitals of the carbon atom share one electron each with the four hydrogen atoms to make a methane molecule. 09_162 lone pair sp 3 sp 3 N H1s H1s sp 3 sp 3 H1s The nitrogen atom in ammonia is sp3 hybridized. A sigma () bond centers along the internuclear axis. A pi () bond occupies the space above and below the internuclear axis. H H C C H H 09_164 2p Hybridization 2p sp 2 E 2s p Orbitals in an isolated carbon atom Carbon orbitals in ethylene An orbital energy-level diagram for sp2 hybridization. 09_165 p orbital sp2 orbital sp2 orbital sp2 orbital In sp2 hybridization one p orbital remains unchanged and lies perpendicular to the plane of the hybrid. 09_166 H1 s H1 s sp 2 sp 2 C C sp 2 H1 s sp 2 sp 2 sp 2 H1 s The shared electron pair of in ethylene occupies the region directly between the atoms to form a sigma () bond. sp2 hybrid orbital • three effective electron pairs. • trigonal planar geometry. • 120 o bond angle. 09_167 p orbital p orbital C C pi bond sigma bond A carbon-carbon double bond consists of a bond and a bond. The bond is formed from unhybridized p orbitals in the space above and below the bond. Pi and Sigma Bonds bonds consist of an electron pair shared in the area centered between the atoms. bonds occupy the space above and below a line joining the atoms. Pi and Sigma Bonds bonds allow rotation. bonds do not allow rotation. 09_168 sp 2 H 1s sp 2 sp 2 sp 2 H 1s C C 2p sp 2 sp 2 (a) H H (b) H C H C The orbitals used to form the bonds in ethylene. 09_169 z z z z z Hybridization x y s x y px x y x y gives a linear arrangement 180° x y Two sp orbitals are formed when one s and one p orbital are hybridized. They are oriented at 180o to each other. sigma bond (1 pair of electrons) 09_174 O pi bond (1 pair of electrons) C O pi bond (1 pair of electrons) (a) O C O (b) The hybrid orbitals in the CO2 molecule. sp hybrid orbital • two effective electron pairs. • linear geometry. • 180 o bond angle. 09_175 p sp N sp p (a) lone pair sigma bond lone pair N sp N sp sp sp (b) N N (c) The nitrogen molecule forms a triple bond -- one and two bonds. dsp3 hybrid orbitals • • • • five effective electron pairs. trigonal bipyramidal geometry. 90 o and 120 o bond angles. hybrid orbitals are not all equivalent as in the other types of hybridization. • Phosphorus pentachloride d2sp3 hybrid orbitals • • • • six effective electron pairs. octahedral geometry. 90 o bond angles. Sulfur hexafluoride. 09_179 Number of Effective Pairs Arrangement of Pairs Hybridization Required 180° 2 Linear sp 3 Trigonal planar sp2 120° 109.5° 4 Tetrahedral sp3 90° 5 Trigonal bipyramidal dsp3 120° 90° 6 Octahedral d2sp3 90° The relationship of the number of effective pairs, their spatial arrangement, and the hybrid orbitals. The Localized Electron Model - Draw the Lewis structure(s) - Determine the arrangement of electron pairs (VSEPR model). - Specify the necessary hybrid orbitals. Deficiencies of the LEM Model • Does not adequately explain resonance. • Does not work for odd-electron molecules and ions. • Assumes that all electrons are localized about an atom. • Gives no direct information about bond energies. Molecular Orbitals (MO) Analagous to atomic orbitals for atoms, MOs are the quantum mechanical solutions to the organization of valence electrons in molecules. Electrons are considered to be delocalized over the entire molecule. Types of MOs bonding: lower in energy than the atomic orbitals from which it is composed. antibonding: higher in energy than the atomic orbitals from which it is composed. 09_556 Energy diagram Electron probability distribution HA H2 HB MO2 E 1s A + + + 1s B MO1 (a) + (b) The molecular orbital energy diagram for the H2 molecule and the MO1 and MO2 orbitals formed. MO1 = 1s and MO2 = 1s*. Bond Order (BO) Difference between the number of bonding electrons and number of antibonding electrons divided by two. # bonding electrons # antibonding electrons BO = 2 Larger bond order means greater bond strength! 09_195 B2 C2 N2 O2 2p* 2p* 2p* 2p* 2p 2p 2p 2p 2s* 2s* 2s 2s F2 E Magnetism Para– magnetic Dia– magnetic Dia– magnetic Para– magnetic Dia– magnetic Bond order 1 2 3 2 1 Observed bond dissociation energy (kJ/mol) 290 620 942 495 154 Observed bond length (pm) 159 131 110 121 143 The molecular orbital energy-level diagrams, bond orders, bond energies, and bond lengths for diatomic molecules. In order to participate in MOs, atomic orbitals must overlap in space. (Therefore, only valence orbitals of atoms contribute significantly to MOs.) 09_187 Li Li 1s 1s 2s 2s The relative size of the lithium 1s and 2s orbitals. The 1s orbital can be considered to be localized and do not participate in bonding. 09_189 B B (a) (b) (c) (d) The boron molecule will form one and two bonds. 09_190 *2p Antibonding 2px 2px (a) 2p Bonding *2p Antibonding 2py 2p 2py (b) Bonding The two p orbitals that overlap head on make two molecular orbitals -- one bonding and one antibonding. The two p orbitals that lie parallel overlap to produce two molecular orbitals, one bonding and one antibonding. Paramagnetism - unpaired electrons - attracted to induced magnetic field - much stronger than diamagnetism - B2 & O2 09_195 B2 C2 N2 O2 2p* 2p* 2p* 2p* 2p 2p 2p 2p 2s* 2s* 2s 2s F2 E Magnetism Para– magnetic Dia– magnetic Dia– magnetic Para– magnetic Dia– magnetic Bond order 1 2 3 2 1 Observed bond dissociation energy (kJ/mol) 290 620 942 495 154 Observed bond length (pm) 159 131 110 121 143 The molecular orbital energy-level diagrams, bond orders, bond energies, and bond lengths for diatomic molecules. Diamagnetism - paired electrons - repelled from induced magnetic field - much weaker than paramagnetism - C2 , N2 , & F2 . 09_193 Balance Glass tubing Sample tube Electromagnet Apparatus used to measure the paramagnetism of a sample. A paramagnetic sample will appear heavier when the electromagnet is turned on. 09_198 H atom HF molecule F atom * 1s E 2p A partial molecular orbital energy-level diagram for the HF molecule. Bond order is 1 -- a single bond. 09_199 H nucleus F nucleus The electron probability distribution in the bonding molecular orbital of the HF molecule. Pi and Sigma Bonds bonds in a molecule are described as being localized. bonds are considered to be delocalized over the entire molecule. NO2 Molecule Draw the Lewis Structure, determine the parent geometry, the actual geometry, and the approximate bond angle. NO+ ION Draw the Lewis Structure, draw the molecular orbital energy-level diagram, determine the bond order, and the type of magnetism for the NO+ ion. 09_202 sp 2 H C H H C sp 2 C C C H H H1s C H The bonding system in the benzene molecule. 09_203 H H H H (a) H H H H H H H H (b) The molecular orbital system in benzene. The electrons in the orbitals are delocalized over the ring of carbon atoms. Outcomes of MO Model 1. As bond order increases, bond energy increases and bond length decreases. 2. Bond order is not absolutely associated with a particular bond energy. 3. N2 has a triple bond, and a correspondingly high bond energy. 4. O2 is paramagnetic. This is predicted by the MO model, not by the LE model, which predicts diamagnetism. Combining LE and MO Models bonds can be described as being localized. bonding must be treated as being delocalized.