LecCh7ProteinStructure
advertisement

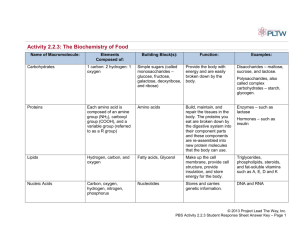
Protein Structure Prediction Why do we want to know protein structure? Classification Functional Prediction What is protein structure? Primary - chains of amino acids Secondary - interaction between groups of amino acids Tertiary - the organization in three dimensions of all the atoms in a polypeptide Quaternary - the conformation assumed by a multimeric protein Primary Structure Proteins are chains of amino acids joined by peptide bonds Polypeptide chain The structure of two amid acids The N-C-C sequence is repeated throughout the protein, forming the backbone The bonds on each side of the C atom are free to rotate within spatial constrains, the angles of these bonds determine the conformation of the protein backbone The R side chains also play an important structural role Secondary Structure Interactions that occur between the C=O and N-H groups on amino acids Much of the protein core comprises helices and sheets, folded into a threedimensional configuration: - regular patterns of H bonds are formed between neighboring amino acids the amino acids have similar angles the formation of these structures neutralizes the polar groups on each amino acid the secondary structures are tightly packed in a hydrophobic environment Each R side group has a limited volume to occupy and a limited number of interactions with other R side groups helix sheet Secondary Structure helix sheet Secondary Structure Other Secondary structure elements (no standardized classification) - random coil - loop - others (e.g. 310 helix, -hairpin, paperclip) Super-secondary structure - In addition to secondary structure elements that apply to all proteins (e.g. helix, sheet) there are some simple structural motifs in some proteins - These super-secondary structures (e.g. transmembrane domains, coiled coils, helix-turn-helix, signal peptides) can give important hints about protein function Classification Structural classification of proteins (SCOP) Class 1: mainly alpha Class 3: alpha/beta Class 2: mainly beta Class 4: few secondary structures More Classification Alternative SCOP Class : only helices Class : antiparallel sheets Class / : mainly sheets with intervening helices Class + : mainly segregated helices with antiparallel sheets Membrane structure: hydrophobic helices with membrane bilayers Multidomain: contain more than one class Protein Structure Review Q: If we have all the Psi and Phi angles in a protein, do we then have enough information to describe the 3-D structure? A: No, because the detailed packing of the amino acid side chains is not revealed from this information. However, the Psi and Phi angles do determine the entire secondary structure of a protein Tertiary structure Secondary-Structure Prediction Programs * PSI-pred * JPRED Consensus prediction (includes many of the methods given below) * DSC * PREDATOR * PHD * ZPRED * nnPredict * BMERC PSA * SSP Tertiary Structure The tertiary structure describes the organization in three dimensions of all the atoms in the polypeptide The tertiary structure is determined by a combination of different types of bonding (covalent bonds, ionic bonds, h-bonding, hydrophobic interactions, Van der Waal’s forces) between the side chains Many of these bonds are very week and easy to break, but hundreds or thousands working together give the protein structure great stability If a protein consists of only one polypeptide chain, this level then describes the complete structure Tertiary Structure Proteins can be divided into two general classes based on their tertiary structure: - Fibrous proteins have elongated structure with the polypeptide chains arranged in long strands. This class of proteins serves as major structural component of cells Examples: silk, keratin, collagen - Globular proteins have more compact, often irregular structures. This class of proteins includes most enzymes and most proteins involved in gene expression and regulation Quaternary Structures The quaternary structure defines the conformation assumed by a multimeric protein. The individual polypeptide chains that make up a multimeric protein are often referred to as protein subunits. Subunits are joined by ionic, H and hydrophobic interactions Example: Haemoglobin (4 subunits) Structure Displays Common displays are (among others) cartoon, spacefill, and backbone cartoon spacefill backbone Software RasMol Cn3D Jmol (Chime) Classic Approach to Determining Structure? Determine biochemical and cellular role of protein Experimentally determine 3D structure Infer function, mechanism of action Purify protein Clone cDNA encoding protein Obtain protein By expression Structural Genomics Approach? genomic DNA sequences predict proteincoding genes Obtain protein by expression Obtain protein In silico Experimentally determine 3D structure Predict 3D structure homology searches (PSI-BLAST) Determine biochemical and cellular role of protein Sources of Protein Structure Information? 3-D macromolecular structures stored in databases The most important database: the Protein Data Bank (PDB) The PDB is maintained by the Research Collaboratory for Structural Bioinformatics (RCSB) and can be accessed at three different sites (plus a number of mirror sites outside the USA): - http://rcsb.rutgers.edu/pdb (Rutgers University) - http://www.rcsb.org/pdb/ (San Diego Supercomputer Center) - http://tcsb.nist.gov/pdb/ (National Institute for Standards and Technology) It is the very first “bioinformatics” database ever build Structural Prediction Computational Modeling Researches have been working for decades to develop procedures for predicting protein structure that are not so time consuming and not hindered by size and solubility constrains. As protein sequences are encoded in DNA, in principle, it should therefore be possible to translate a gene sequence into an amino acid sequence, and to predict the three-dimensional structure of the resulting chain from this amino acid sequence Computational Modeling How to predict the protein structure? Ab initio prediction of protein structure from sequence: not yet. Problem: the information contained in protein structures lies essentially in the conformational torsion angles. Even if we only assume that every amino-acid residue has three such torsion angles, and that each of these three can only assume one of three "ideal" values (e.g., 60, 180 and -60 degrees), this still leaves us with 27 possible conformations per residue. For a typical 200-amino acid protein, this would give 27200 (roughly 1.87 x 10286) possible conformations! Q: Can’t we just generate all these conformations, calculate their energy and see which conformation has the lowest energy? If we were able to evaluate 109 conformations per second, this would still keep us busy 4 x 10259 times the current age of the universe There are optimized ab initio prediction algorithms available as well as fold recognition algorithms that use threading (compares protein folds with know fold structures from databases), but the results are still very poor Homology Modeling Homology (comparative) modeling attempts to predict structure on the strength of a protein’s sequence similarity to another protein of known structure Basic idea: a significant alignment of the query sequence with a target sequence from PDB is evidence that the query sequence has a similar 3-D structure (current threshold ~ 40% sequence identity). Then multiple sequence alignment and pattern analysis can be used to predict the structure of the protein Computational modeling: summary Partial or full sequences predicted through gene finding Similarity search against proteins in PDB Find structures that have a significant level of structural similarity (but not necessarily significant sequence similarity) Alignment can be used to position the amino acids of the query sequence in the same approximate 3-D structure If member of a family with a predicted structural fold, multiple alignment can be used for structural modeling How do we do this? Structural analyses in the lab (X-ray crystallography, NMR) Infer structural information (e.g. presence of small amino acid motifs; spacing and arrangement of amino acids; certain typical amino acid combinations associated with certain types of secondary structure) can provide clues as to the presence of active sites and regions of secondary structure 3D Comparative Modeling Profile Methods - match sequences to folds by describing each fold in terms of the environment of each residue in the structure Threading Methods - match sequences to structure by considering pairwise interactions for each residue, rather than averaging them into an environmental class HMM Methods - the equivalent state corresponds to one structurally aligned position in a structural fold, including gaps Structural HMM