PowerPoint 프레젠테이션 - Springer Static Content Server
advertisement
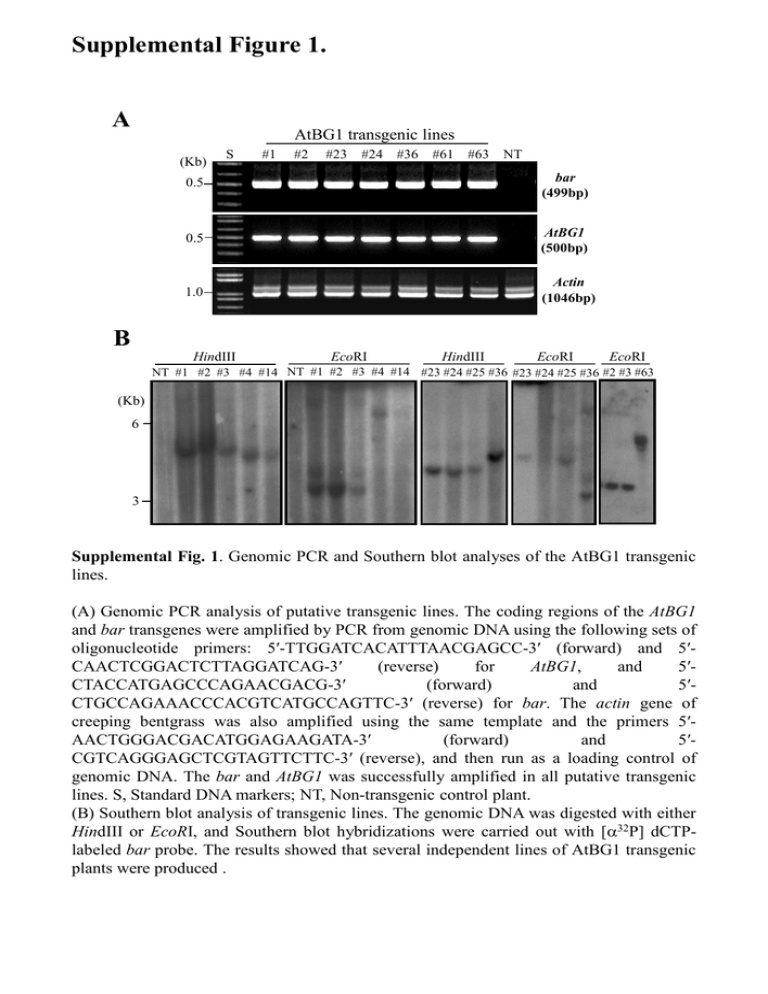
Supplemental Figure 1. A AtBG1 transgenic lines (Kb) S #1 #2 #23 #24 #36 #61 #63 NT 0.5 bar (499bp) 0.5 AtBG1 (500bp) 1.0 Actin (1046bp) B HindIII EcoRI HindIII EcoRI EcoRI NT #1 #2 #3 #4 #14 NT #1 #2 #3 #4 #14 #23 #24 #25 #36 #23 #24 #25 #36 #2 #3 #63 (Kb) 6 3 Supplemental Fig. 1. Genomic PCR and Southern blot analyses of the AtBG1 transgenic lines. (A) Genomic PCR analysis of putative transgenic lines. The coding regions of the AtBG1 and bar transgenes were amplified by PCR from genomic DNA using the following sets of oligonucleotide primers: 5′-TTGGATCACATTTAACGAGCC-3′ (forward) and 5′CAACTCGGACTCTTAGGATCAG-3′ (reverse) for AtBG1, and 5′CTACCATGAGCCCAGAACGACG-3′ (forward) and 5′CTGCCAGAAACCCACGTCATGCCAGTTC-3′ (reverse) for bar. The actin gene of creeping bentgrass was also amplified using the same template and the primers 5′AACTGGGACGACATGGAGAAGATA-3′ (forward) and 5′CGTCAGGGAGCTCGTAGTTCTTC-3′ (reverse), and then run as a loading control of genomic DNA. The bar and AtBG1 was successfully amplified in all putative transgenic lines. S, Standard DNA markers; NT, Non-transgenic control plant. (B) Southern blot analysis of transgenic lines. The genomic DNA was digested with either HindIII or EcoRI, and Southern blot hybridizations were carried out with [32P] dCTPlabeled bar probe. The results showed that several independent lines of AtBG1 transgenic plants were produced . Supplemental Figure 2. NT #1 #2 #23 #24 Supplemental Fig. 2. Apparent phenotypes of representative AtBG1 transgenic plants. Fully-grown AtBG1 transgenic plants and non-transgenic control plants (NT) were shown. Supplemental Figure 3. TATTATCATT GCAAGTGATA CCATTGCCAA TTCTTTTTAT AGCTGGTATG GTAAAGTATG GTTTTTCCTT CCAAAAGATC ACCCCTGGTG AGCTCAGCGC TGCTTGACAG CGGACGTTTT CTGGATTTTG ACATACTAAG BAR CGAGTGACGA AAATATTCAG CATCCAACAT GAAAGAAAAT GGCAGGCCAG AGCTGAATGG GACACATCTC AACTACATAC AAATGCTAAC AGCAAATTTG GACAGCCATT TAATGTACTG GTTTTAGGAA GGTTTCTTAT 35S CGTCCACACC TTGGTTGCAA GAAGGAGTTA AAGAAAAGGG ATAGCCAATA CCGAAGCTCT CGGAAGACAG CTATATGTTT TAGGAGTAGA AACAAGACGC CCACAGCGAG AATTAACGCC TTAGAAATTT ATGCTCAACA AtBG1 AGAAAAGAAA CAGGAAAACT ATTGAGATCT AGATAACGAA GCTTTGTCTG TGTCATGGAT ATCCGTTCCA GACAAGGAAA GTAGAATAAT GAGGGGTAGC TGACGAGATC GAATTAATTC TATTGATAGA CATGAGCG ATTTTAGTTG TTCAAGTTGC GAGTCAATTC GGACGCAATT CCCGTCGTAT GTGTATGCAT AAAACCCTGA ATTCCCGCAA TCAATAAACT ACAAGCACAA GACAACTTAA GGGGGATCTG AGTATTTTAC Ubiquitin-P 35S CCACGTGTGA AAAGTTTCTT TGAATTACGT TTTTTATGAT GCGCAAACTA TCTAAACAAT ATTCACCTGT TTAGATATAT TACTGTGCAG AGAGAGAACA AGATATGCAT TCTCCTCGTA ATTACAGGTG AAGATTGAAT TAAGCATGTA TAGAGTCCCG GGATAAATTA GCATGTTCAC TTTTTACTAC TTGAACACTA CTCACAGTAT AAAAGTTGTC GTCCTCATTC ATATTATTAG CGACCATAGT ACGGTTGCAC TCAATCGTAT ATTCATGGCA AGAATTTCTG GCACGCCCCT TGTAGTAAGT AATTCACGCT GATTGTAAAC TCCCTTGTAC TAACACATTG GATTTTAGTA AAATACAAAT 35S poly A tail Intron-GUS GCAGGG ACCAGTTCGA CCTGTTGCCG ATAATTAACA CAATTATACA TCGCGCGCGG CTATCATTGA TTTATCCGTC CTCATATTGA ACAGCCACAT AATACTAAAA TCATAATTTT CTGAGGTGAT NOS terminator AGGCAAACAA ATTTCCCCGA GTCTTGCGAT TGTAATGCAT TTTAATACGC TGTCATCTAT AAGAATATCG CTATAAAGGA CATCTTTACA ATACCAAGTT GGAAAAAAAC TTCTGTCCGT GCTAGCCACC TCGTTCAAAC GATTATCATA GACGTTATTT GATAGAAAAC GTTACTAGAT AATGTGAATT TGTCATAAAT TTTACAGATA TGTCTCTCAC ATTGTCTGCC AGCTTCTCAT ACCACCACCA ATTTGGCAAT TAATTTCTGT ATGAGATGGG AAAATATAGC GATGCTAGCA ATGCATAGGT TTGTATAAAC TGCAATCAAG AAAAATATAG AATGCCAGTC CCCGTGAACA Supplemental Fig 3. Determination of the T-DNA insertion site in AtBG1 #23 transgenic line by I-PCR. Inverse PCR (I-PCR) of transgenic plants were performed as previously reported (Ochman et al., 1988) with some modifications. Total genomic DNA was isolated from the leaves of greenhouse-grown plants and restriction digests were carried out using 2 μg of genomic DNA. For circularization, restriction fragments were self-ligated overnight and 50 ng of circularized DNA were used for the first round I-PCR. The DNA products from the first round PCR reactions were used for the second round PCR and the PCR products were fractionated on a 0.8% agarose gel. The purified PCR products were cloned into pGEM-T vector (Promega) and sequencing analysis was performed. The flanking sequences of TDNA were analysed using BLAST in NCBI site, and the insertion of T-DNA was found to be a non-coding region. Non-coding region was shown in Black, and CaMV 35S polyA and NOS terminator of T-DNA region in the #23 transgenic plant were shown in red and blue, respectively. The T-DNA region of the binary vector plasmid pCAMBIA3301 harbouring AtBG1 was shown in the middle of sequence. Ochman H, Gerber AS, Hartl DL (1988) Genetic applications of an inverse polymerase chain reaction. Genetics 120:621-623 Supplemental Figure 4. TCTGCGTCGC AGGAATGCCG TTGGCCGTTA AGCTCCACGA GAGATATATG ATACAGTCAC CCGGTGATAT GAACGAAGAA GACGTTTTTA GGATTTTGGT ATACTAAGGG CGTCGATCAC CCATTGCACC TGTGTGGGTG ATCTGTGAAA CATCCTTATC ATTAATTATT GGTTGCAAAT TATTGTGGTG ATGTACTGAA TTTAGGAATT TTTCTTATAT BAR 35S AGCTCGCATG AAACTACAAG AAAACCTGCT AGAATACATA TAATGTCTGA GCACATGGGC ATTAATTGGA TAAACAAATT TTAACGCCGA AGAAATCTTA GCTCAACACA AtBG1 TTCATCTTGT AGTGTGAGAG ACCCTGGTTG TGCATGGATC ACTCTCAAGT AGCTGAGTTA TGCAGACAGA GACGCTTAGA ATTAATTCGG TTGATAGAAG TGAGCGAATC GGGTCATAAG GAAGCACTGC ACCTTAGGCT GTTAGGCAGG CTCTTATATA TGCACGGCGA GATATGACAA TATGCCAATC CAATATAAAT TTGGATTAAA GCATTCGACC ATGCATAGCT TAACCATCAC GAGTTTAGTG CAACTTAATA ACACATTGCG GGGATCTGGA TTTTAGTACT TATTTTACAA ATACAAATAC GAATTC 35S poly A tail Ubiquitin-P 35S Intron-GUS CGATGATTAT GCATGACGTT ACGCGATAGA CTATGTTACT ATCGAATGTG AGGATGTCAT TACATTTACA AGTTTGTCTC AAACATTGTC CCGTAGCTTC GTGTATACCT CATATAATTT ATTTATGAGA AAACAAAATA AGATGATGCT AATTATGCAT AAATTTGTAT GATATGCAAT TCACAAAAAT TGCCAATGCC TCATCCCGTG GTCAAAGTAA NOS terminator ACTAGTGATT ACGTTAAGCA TGATTAGAGT ACTAGGATAA CAATGCATGT CTGTTTTTTA ATATTTGAAC GCAGCTCACA AACAAAAAGT GCATGTCCTC CGTAATATTA GAATCCTGTT TGTAATAATT CCCGCAATTA ATTATCGCGC TCACCTATCA CTACTTTATC ACTACTCATA GTATACAGCC TGTCAATACT ATTCTCATAA TTAGCTGAGG GCCGGTCTTG AACATGTAAT TACATTTAAT GCGGTGTCAT TTGAAAGAAT CGTCCTATAA TTGACATCTT ACATATACCA AAAAGGAAAA TTTTTTCTGT TTATTACCCG CTGTTGAATT TGGGTTTTTA TAGCGCGCAA AGCATCTAAA AGGTATTCAC AAATTTAGAT CAAGTACTGT ATAGAGAGAG AGTCAGATAT AACATCTCCT GATAAAATCG Supplemental Fig. 4. Determination of T-DNA insertion site in AtBG1 #2 transgenic line by I-PCR. Inverse PCR on #2 transgenic plant was performed using the same methods described in Supplemental Figure 3. The T-DNA was integrated into a non-coding region. The noncoding region was shown in Black, and CaMV 35S polyA and NOS terminator of T-DNA region in the #2 transgenic plant were shown in red and blue, respectively. The T-DNA region of the binary vector plasmid pCAMBIA3301 harbouring AtBG1 was shown in the middle of sequence.







