Ab 1-16 Cu 2+ /Zn 2+
advertisement

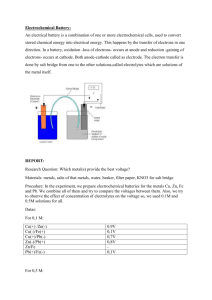
Copper and Zinc Coordination mode in b-Amyloid Peptides A XAS and ab initio study V. Minicozzi Phys. Dept.- University of Rome Tor Vergata ABR2008, 10-11 Aprile 2008, Roma Biophysics Group in Rome “Tor Vergata” Experiments CNR- Trento M. Dalla Serra C. Potrich S. Morante G. C. Rossi V. Minicozzi F. Stellato S. Alleva A. Maiorana Calculations DESY -Berlin K. Jansen N. Christian EMBL-DESY-Hamburg Computations have been performed at W. Meyer-Glauche Fermi BEN Altix Summary • Ab peptide • XAS results • ab-initio simulations • Conclusions and outlook Alzheimer Disease (AD) AD brains show two lesions Neurofibrillar Tangles 1- Amyloid Plaques: Extracellular peptide deposits of Ab almost spherical with a 10-100 mm diameter 2- Neurofibrillar Tangles: Intracellular abnormal elicoidal fibers mainly composed by tau protein These two lesions can occur independently of each other Neuron Amyloid Plaques Amyloid b-peptide •- & -secretases cleavage nonpathological peptide P3 • Ab is derived from proteolitic cleavage of APP protein (Amyloid Precursor Protein). •APP: 770 trans-membrane protein coded in chromosome 21 •b- & -secretases cleavage pathological peptides Ab1-40, Ab1-42 APP -secretase P3 17 40-42 -secretase APP b-secretase Ab 1 40-42 Cu2+ Cu2+ EPR NMR J. Danielsson et al. (2007) FEBS 274:46 A.K. Tickler et al. (2005) JBC 280:13355 Zn2+ Zn2+ NMR NMR S. Zirah et al. (2006) JBC 281:2151 Syme & Viles (2006) BBA 1764:246 Experiments on… DAEFRHDSGY EVHHQKLVFF AEDVGSNKGA IIGLMVGGVV Cu2+/Zn2+- Ab1-16 minimal fragment containing His6, His13, and His14, suggested to be involved in metal binding Cu2+/Zn2+- Ab17-40 complementary sequence where none of these His’s is present Cu2+/Zn2+- besides the presence of these three His’s, a long hydrophobic region believed to be relevant in the aggregation process Ab1-28 Cu2+/Zn2+- Ab5-23 the N-terminal region of the Ab-peptide can play any role in the metal binding process? • Stellato et al., Eur Biophys J (2006) 35: 340 • Minicozzi et al., (2008) J Biol Chem in press EXAFS Cu2+/Zn2+-Ab17-40 = Cu2+/Zn2+- buffer Cu2+-Ab1-16 = Cu2+- Ab1-28 = Cu2+-Ab1-40 Cu2+-Ab5-23 ≠ Cu2+-Ab1-16 Zn2+-Ab1-16 = Zn2+-Ab1-28 = Zn2+-Ab5-23 = Zn2+-Ab1-40 Cu-Ab1-16 EXAFS |FT| Data Fit 0 1 2 3 •3 Histidines •1 Tyrosine •1 O 4 r(Å) 5 Cu-Ab5-23 EXAFS |FT| Data Fit 0 1 •2 •1 •1 •1 2 Histidines N-term Tyrosine O 3 4 r(Å) 5 Zn-Ab1-16 EXAFS |FT| Data Fit 0 1 2 •4 Histidines •1 O 3 4 r(Å) 5 XAS Conclusions • Metal binding site lies within the first 16 aminoacids • Cu2+ and Zn2+ have different binding geometry • Zn2+-Ab less rigid geometry, sensitive to solution condition • Cu2+-Ab very stable binding mode • Zn2+-Ab inter-molecular binding suggests aggregation • Cu2+-Ab intra-molecular binding Questions: 1. precise location of metal binding site along the sequence 2. different zinc and copper role in aggregation processes A promising tool is ab initio molecular dynamics Car-Parrinello Molecular Dynamics simulations (CP-MD) Classical MD atoms move in the chosen force field ab-initio MD electrons are active quantum mechanical DOF CP method is based on DFT theory ℒ i r 2 i 1 M I R I2 E i , RI ij [ i j ij ] 2 I ij Enforcing the orthonormality of KS wave functions Nuclei move experiencing both the force due to electrons, EDFT , and the force due to electrostatic nuclear interaction, EN Fictitious dynamics for electrons i ⇨ electronic degrees of freedom i E ij j i j parallel version of Quantum-ESPRESSO package ⇨ CP-MD • Vanderbilt ultrasoft pseudopotentials • Perdew-Burke-Ernzerhof (PBE) exchange-correlation (xc) functional Ab-peptide systems Classical MD simulations of Cu+2-Ab1-16 in water: - Cu+2 bounded to His6, Tyr10, His13, His14 - Cu+2 bounded to Nterm, His6, His13, His14 S2 Cu+2(D1-2-3-4-5-H6-7-8-9-Y10-E11-12-H13-H14-cap) + 180 H2O Classical MD simulations of Zn+2-Ab1-16 in water: comes from classical MD of Cu+2-Ab1-16 with Cu+2 bound to His6, Tyr10, His13, His14 - Zn+2 bounded to His6, Tyr10, His13, His14 - Zn+2 bounded to 4 Histidines 2 Ab1-16 CP-MD simulations of Cu complexes S3 Cu+2(D1-2-3-4-5-H6-7-8-9-Y10-E11-12-H13-H14-cap) + 158 H2O +2(D -2-E -4-5-H -cap cap-H -H -cap) + 125 H O S1comes Cu 1 3 MD of Cu 6 +2-Ab1-16 with13 2 6, His13, His14 from classical Cu+2 14 bound to Nterm, His S2 Cu+2(D1-2-3-4-5-H6-7-8-9-Y10-E11-12-H13-H14-cap) + 180 H2O S3 Cu+2(D1-2-3-4-5-H6-7-8-9-Y10-E11-12-H13-H14-cap) + 158 H2O S1 494 atoms and 1369 electrons S2 703 atoms and 1951 electrons S3 628 atoms and 1776 electrons S1: Distances from Cu2+ S2: Distances from Cu2+ Conclusions and Outlook • We can discriminate via ab-initio simulations among the structural models extracted from XAS experiments • QM/MM simulations of the whole hydrated Ab peptide based on structural XAS data expecially relevant for Zn+2 structures where pairs of peptides are involved • New XAS experiments with Aluminium and Zinc Thanks for your attention! XANES Cu2+-Aβ1-16 = Cu2+-Aβ1-28 = Cu2+-Aβ1-40 ≠ Cu2+-Aβ5-23 ≠ Cu2+-Aβ17-40 Zn2+-Aβ1-16 = Zn2+-Aβ1-28 ≠ Zn2+-Aβ1-40 ≠ Zn2+-Aβ5-23 ≠ Zn2+-Aβ17-40 Feasibility studies for S1 system Platform CPU hours for 104 steps Fermi1 in a 16 node configuration 1650 BEN in a 16 node configuration 1300 on S1 Altix scaling Altix in a 16 node configuration 300 psbased at 300 K GHz - Fermi1 Linux-clusters (E. Fermi Institute, Rome -3.6 Italy) on 1.7 Pentium IV processors 64 nodes ~ 1 month - BEN Linux-cluster (ECT∗ Institute, Trento - Italy) based on Intel/XeonV.Minicozzi, et al. (2008) International Journal of Quantum 2.8 GHz processors Chemistry in press - ALTIX 4700 (LRZ, Munich - Germany) based on Intel Itanium2 Madison 9M 1.6 GHz processors S1: Dihedral angle Nδ(H6)–Cu–Ne(H13)–Nδ(H14) as a function of the CP simulation time