Document
advertisement
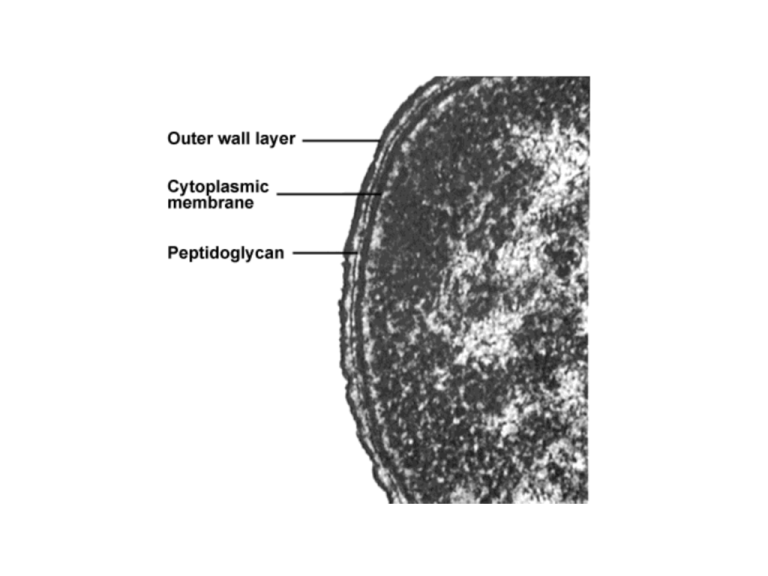
Microscopy • Compound Light Microscope – Objective lens = real image – Eye piece = virtual image – Magnification – Condenser lens and iris diaphragm – Other terms • Resolving power (resolution) • Refractive index and immersion oil • Refractive index of bacteria ~ water (so invisible) Microscopy cont. Bright Field • Stains, Gram, spore, flagella, Sudan black etc. • Cell surface is negative – therefore stains positive • Simple stains • Differential stains Phase Contrast and Dark Field • Phase: amplifies the slight difference in the refractive index and converts the difference into contrast. • Dark:special condenser –only light from specimen enters objective Fluorescence • Uv or halogen light source • Illuminate from above • Use different filters to select for different wavelengths • Cyanobacteria glow red =chlorophyll and other pigments =autofluorescence • Stains, ribosomal RNA probes, DAPI etc. Microscopy cont. 3-D imaging • Differential Interference Contrast (plane polarized) • Atomic Force (repulsive atomic forces) • Confocal Scanning Laser Microscopy (laser light) Electron Microscopy TEM Electron gun = e- which is the illumination Get REALLY short wavelengths = greater resolution Need to make thin sections SEM Morphology • • Cell size, why be small? Morphology cont. • Unicellular • Rods or bacillus, vibrio spirullum • Cocci (chains, diplococci, grapes) • Filaments, sometimes filaments can be deceptive. To the naked eye you think they are filaments but under the microscope short rods stick together as filaments (PIC from Yellowstone) • Multicellular, like actinomycetes, mycelium (PIC deep-sea Actinomycetes) • Oscillatoria makes a trichome (PIC) • Shealthed and filamentous Division Binary, septum produced along transverse axis DNA replication occurs before septum formation Budding, less common in prokaryotes (some Archaea like Sulfurococcus) Fragmentation, actinomycetes do this, filament fragments to form unicellular rods Fine Structure, Composition and Function Cell membranes Cell membranes, the ultimate barrier between cytoplasm and external environment, gases & water (small uncharged molecules pass through) diffuse readily, ions do not Bacteria, ester linked Archaea, glycerol linked ethers (thermophilic microbes >>> tetraethers) Fine Structure, Composition and Function Bilayer Sterols vs hopanoids Bacterial, eukaryal and archaeal membrane lipids Ester link Ether link Side chains are fatty acids Isoprene side chain = Archaea Structure of Archaeal membranes Note, monolayer Structure and function-membrane transport proteins • Transport proteins Group translocation • Substrate chemically altered Cell Walls • Bacteria, almost all have peptidoglycan (murein), over 100 different peptidoglycan structures , differences are based on the amino acids and how they cross link • N-acetylglucosamine • N-acetylmuramic acid • Lysozyme sensitive Archael cell walls • Archaea have pseudopeptidoglycan contains Nacetylglucosamine (like Bacteria) and Nacetytalosaminuronic acid (unlike Bacteria which have of N-acetylmuramic acid) Bacterial Cell walls • BACTERIA – Gram + 40-80% is peptidoglucan • teichoic acids, polyol phospate polymers – Gram + • No teichoic acids • Only one layer of peptidoglycan, (5% of cell wall weight) • Outer membrane similar to cytoplasmic membrane, lipids, proteins but also polysachharides • LPS or lipopolysaccharide layer (lipid A= endotoxin, bubonic plague, typhoid fever etc.) • Proteins like porins Omp C and Omp F Antibiotics and cell walls • Antibiotics and bacterial cell walls Function to inhibit production of enzymes that make peptidoglycan (eg Penicillin) • Why are Gram + more sensitive than Gram -? LPS-Gram –ive Bacteria • • LPS or lipopolysaccharide layer (lipid A= endotoxin, bubonic plague, typhoid fever etc.) Proteins like porins Omp C and Omp F Capsules • Protective outer layer made up of polysaccharides, some polypeptides • Often house the virulence factors eg. Steptococcus pneumoniae Protein layers, -S-layer, sheaths • S-layer, perhaps involved in mineral precipitation? • Very fragile • Sheaths-complex composition, important to many iron oxidizing bacteria Fine Structure, Composition and Function • DNA , concentrates in an area in cytoplasm= nucleoid In general: 4 X 106 Mbp Plasmids (carry important functions like metal resistance, naphthalene degradation, antibiotic resistance; also may be important for lateral gene transfer) Transcription Transduction, tranformation, conjucation • Ribosomes, 30S + 50S = 70S (‘cause Svedberg unit not directly related to molecular mass, rather density!) Antibiotic sensitivity translation Genome size Flagella • Filament – Flagellin – Hollow; self assembly – wavelength • Powered by PMF • 1000 protons/rotation! • FAST Motility Polar and peritrichous flagella Taxis Cell surface structures • Fimbria (-ae) – Structurally like flagella, but shorter – Various functions including adhesion • Pilus (-i) – Longer than fimbriae – Functions include conjugation • S-layer • Capsule or slime layer – Collectively called glycocalyx – Avoidance of phagocytosis, dessication Storage materials and inclusions • Carbon-storage polymers – Poly--OH-alcanoate (PHA) – Poly--OH-butyrate (PHB)- lipid – Glycogen, Starch (alfa 14 glucose linkages) • Polyphosphate • Sulfur • Cyanophycin , (nitrogen polymer in cyanobacteria) • Magnetite (Fe3O4 ) • Gas vesicles (membrane structures found in a wide diversity of microbes, many aquatic microbes for buoyancy) (Antartica, Jim Staley, U Washington) Gas diffuses freely across the membrane Endospores • Resistant to heat, drying, etc. • Survival, not procreation• Spores in amber (25-40 My) • Bacillus and Clostridium Early stages of endospore formation Middle stages of endospore formation Completion of endospore formation See table 3.2 Eukaryotic cells • • • • DNA in nucleus DNA arranged in chromosomes Ribosomes: 80S Organelles – Mitochondrion (-ia) – Chloroplast Mitochondria and chloroplasts are prokaryotes • Contain DNA – Closed, circular • Prokaryotic ribosomes (70S) • Antibiotic sensitivity • Ribosomal phylogeny – Mitochondria are related to proteobacteria – Chloroplasts are related to cyanobacteria