Analyze_AAS_0.4 - QI
advertisement

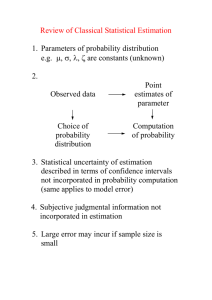
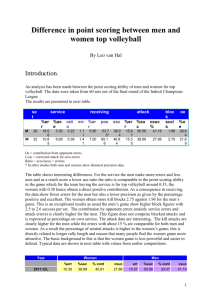
QI-Bench “Analyze” AAS Rev 0.4 QI-Bench “Analyze” Architecture Specification August 14, 2012 Rev 0.4 Required Approvals: Author of this Revision: Andrew J. Buckler System Engineer: Andrew J. Buckler Print Name Signature Date Document Revisions: Revision Revised By Reason for Update Date 0.1 AJ Buckler Initial version June 2011 0.2 AJ Buckler Updates February 4, 2012 0.3 AJ Buckler To close Iteration 1 August 14, 2012 0.4 AJ Buckler To open Iteration 2 August 14, 2012 BBMSC 1 of 15 QI-Bench “Analyze” AAS Rev 0.4 Table of Contents 1. EXECUTIVE SUMMARY ................................................................................................ 3 1.1. PURPOSE AND SCOPE ..................................................... ERROR! BOOKMARK NOT DEFINED. 2. STRUCTURE OF THE APPLICATION ......................................................................... 3 2.1. DATASTORES ......................................................................................................................... 6 2.1.1. Biomarker DB ................................................................................................................ 6 2.1.2. Re-useable Library of Analysis Scripts .......................................................................... 6 2.1.3. Assessment DB ............................................................................................................... 7 2.2. ACTIVITIES ............................................................................................................................ 7 2.3. ASSUMPTIONS ........................................................................................................................ 7 2.4. DEPENDENCIES ...................................................................................................................... 8 3. SYSTEM-LEVEL REQUIREMENTS ............................................................................. 8 3.1. FUNCTIONALITY STAGED FOR FIRST DEVELOPMENT ITERATION ........................................... 8 3.2. PERFORMANCE..................................................................................................................... 12 3.3. QUALITY CONTROL ............................................................................................................. 12 3.4. “SUPER USER” AND SERVICE SUPPORT ................................................................................ 12 3.5. UPGRADE / TRANSITION ....................................................................................................... 13 3.6. SECURITY ............................................................................................................................ 13 3.7. REQUIREMENTS FOR SUBSEQUENT ITERATIONS ................................................................... 14 4. DEPLOYMENT MODEL(S) ........................................................................................... 15 5. IMPLEMENTATION CONSIDERATIONS AND RECOMMENDATIONS FOR TECHNICAL REALIZATION ................................................................................................. 15 BBMSC 2 of 15 QI-Bench “Analyze” AAS Rev 0.4 1. Executive Summary The purpose of QI-Bench is to aggregate evidence relevant to the process of implementing imaging biomarkers to allow sufficient quality and quantity of data are generated to support the responsible use of these new tools in clinical settings. The efficiencies that follow from using this approach could translate into defined processes that can be sustained to develop and refine imaging diagnostic and monitoring tools for the healthcare marketplace to enable sustained progress in improving healthcare outcomes. 1.1. Purpose and Scope Specifically, Analyze is developed to allow users to: Characterize the method relative to intended use. Apply the existing tools and/or extend them. From a technology point of view, Analyze refers to the part of the project most closely associated with the statistical analysis and specifically including the library of reference statistical analysis methods. Its job is to enrich the logical specification with statistical results. 1.2. Terms Used in This Document The following are terms commonly used that may of assistance to the reader. AAS ASD BRIDG caBIG caDSR CAT CBIIT CFSS CIM DAM EAS ECCF EOS ERB EUC IMS KC NCI NIH PIM PSM BBMSC Application Architecture Specification Application Scope Description Biomedical Research Integrated Domain Group Cancer Biomedical Informatics Grid Cancer Data Standards Registry and Repository Composite Architecture Team Center for Biomedical Informatics and Information Technology Conceptual Functional Service Specification Computational Independent Model Domain Analysis Model Enterprise Architecture Specification Enterprise Conformance and Compliance Framework End of Support Enterprise Review Board Enterprise Use-case Issue Management System (Jira) Knowledge Center National Cancer Institute National Institutes of Health Platform Independent Model Platform Specific Model 3 of 15 QI-Bench “Analyze” AAS PMO PMP QA QSR SAIF SDD SIG SUC SME SOA SOW UML UMLS VCDE Rev 0.4 Project Management Office Project Management Plan Quality Assurance FDA’s Quality System Regulation Service Aware Interoperability Framework Software Design Document Service Implementation Guide System Level Use-case Subject Matter Expert Service Oriented Architecture Statement of Work Unified Modeling Language Unified Medical Language System Vocabularies & Common Data Elements 2. Structure of the Application Describe the application in macro and its overall organization Include an executive summary of the overall application workflow (e.g., a concise overall description of the responsibilities of this application, and the roles (if any) that it plays in interaction patterns such as client-server, service-to-service, etc.) Enumeration of the behavioral interfaces that are known to be needed, with a concise description of each Consider representation formalism and the intended audience, not necessarily rigorously expressing the content in UML BBMSC 4 of 15 QI-Bench “Analyze” AAS Rev 0.4 The behavioral model is understood in the context of the following logical information model: BBMSC 5 of 15 QI-Bench “Analyze” AAS Rev 0.4 Analyze is defined as an implementation of the following behavioral model: 2.1. Datastores 2.1.1. Biomarker DB linked concept instances that represent biomarkers 2.1.2. Re-useable Library of Analysis Scripts Module Specification Status Method Comparison Radar plots and related methodology based on readings from multiple methods on data set with ground truth Currently have 3A pilot in R, not yet generalized but straightforward to do so. BBMSC 6 of 15 QI-Bench “Analyze” AAS Module Rev 0.4 Specification Status Plan to refine based on Metrology Workshop results and include case of comparison without truth also. Bias and Linearity According to Metrology Workshop specifications Demonstrate version today that works from summary statistics, e.g., to support meta-analysis. Plan to add analysis of individual reads. Test-retest Reliability According to Metrology Workshop specifications Prototype demonstrated last month. Plan to build real module in next month. Reproducibility (including detailed factor analysis) Accepts as input fractional factorial data of cross-sectional biomarker estimates with range of fixed and random factors, produces mixed effects model Module under development that will support both meta-analysis as well as direct data. Variance Components Assessment Accepts as input longitudinal change data, estimates variance due to various nontreatment factors Module under development to support direct data. 2.1.3. Assessment DB … 2.2. Activities Activity Model View Controller develop analytical method to support testable hypothesis (e.g., in R scripts) perform an analysis for a specific test (repeat across tests considered for group analysis) annotate biomarker db with results for this test perform an analysis to characterize performance of the group annotate biomarker db with results of group statistic any of these activities may be performed on behalf of one or more sponsors by trusted broker so as to protect individual identities and/or perform on sequestered data 2.3. Assumptions In this section please address the following questions: BBMSC 7 of 15 QI-Bench “Analyze” AAS Rev 0.4 Upon what services does this specification depend (underpinning infrastructure, other HL7 services, etc) Are there any key assumptions that are being made? 2.4. Dependencies List of capabilities (aka responsibilities or actions) that the application’s workflow depends on and description on what it does in business terms. Description Doc Title Doc Version <business friendly description> <document title> <document version> 3. System-level Requirements Note that, the normal process of requirements development does not guarantee that adjacent requirements are directly related. In situations where requirements are tightly related or where requirements are to be considered in the context of an upper level requirement, explicit parent-child relationships have been created. These can be identified by the requirement numbering – child requirements have numbers of the form XX.Y indicating the Yth child of requirement XX. The following list of attributes is used: Origin – Identifies the project or Enterprise Use Case that originated the requirement. Comment / TI – Additional information regarding the requirement. This may include information as to how the requirement may be tested (i.e. the test indication). Design Guideline – Used to identify requirements that are to be taken as guidance or are otherwise not testable. In such cases the phrase “Not a testable requirement” will appear. Requirements may, and often do, apply to multiple components. In such cases, the Component attribute will identify all components where the requirement applies (including components that do not apply to this Enterprise Use Case). The Origin of a requirement is intended to identify the program for which the requirement was originally defined. Often this is SUC (System Use Case) but it may be different. 3.1. Functionality Staged for First Development Iteration Model: Requirements placed on input, output, or significant intermediate data used by the application. BBMSC 8 of 15 QI-Bench “Analyze” AAS Rev 0.4 View: Supported views and other GUI requirements. Note: use of figures for screen shots is encouraged as necessary to show an essential feature, but not to show unnecessary implementation detail. Controller: Functional requirements on logical flow. Requirements Origin Activity: Develop analytical method to support testable hypothesis (e.g., in R scripts) EA There are essentially two types of experimental studies supported: SUC One is a correlative analysis where an imaging measure is taken and a coefficient of correlation is computed with another parameter, such as clinical outcome. A second type is where a measure of accuracy is computed, which necessitates a representation of the “ground truth,” whether this be defined in a manner traceable to physical standards or alternatively by a consensus of “experts” where physical traceability is either not possible or feasible. This workflow is utilized in the latter case. Definition of what constitutes “ground truth” for the data set is established and has been checked as to its suitability for the experimental objective it will support. Support hypothesis testing of the form given in decision trees and claims for context of use Customize the analysis tool. SUC Analyze data with known statistical properties to determine whether custom statistical tools are producing valid results. Customize built-in statistical methods SUC Add the measures, summary statistics, outlier analyses, plot types, and other statistical methods needed for the specific study design. Load statistical methods into analysis tool SUC View Controller SUC SUC SUC SUC SUC SUC Configure presentation of longitudinal data SUC Customize outlier analysis SUC Configure the report generator to tailor the formats of exported data views. The report generator exports data views to files according to a run-time-configured list of the data views that should be included in the report. EA Activity: Perform an analysis for a specific test (repeat across tests considered for group analysis) SUC The technical quality of imaging biomarkers is assessed with respect to the accuracy and precision of the related physical measurement(s). The following comparisons of markups can be made: SUC SUC Analyze statistical variability SUC Measurements of agreement SUC User-defined calculations SUC Correlate tumor change with clinical indicators SUC Calculate regression analysis SUC Calculate factor analysis SUC BBMSC Model 9 of 15 QI-Bench “Analyze” AAS Requirements Rev 0.4 Origin Calculate ANOVA SUC Calculate outliers SUC Provide review capability to user for all calculated items and plots SUC Drill-down interesting cases, e.g., outlying sub-distributions, compare readers on hard tumors, etc. EA Activity: Annotate biomarker db with results for this test EA Activity: Perform an analysis to characterize performance of the group SUC Follow workflow “Analyze an Experimental Run.” SUC Qualitative pairwise algorithm comparison. The goal is to provide a validation assessment of technical and clinical performance via high-throughput computing tasks. Assessment and characterization of variability, minimum detectable change, and other aspects of performance in the intended environment including subject variability associated with the physiological and pathophysiological processes present in the target population – that is, moving beyond the more highly controlled conditions on which the biomarker and its tests may have been initially discovered and developed. The studies are undertaken in part to provide data to support proposed cut-points (i.e., decision thresholds), if imaging results are not reported as a continuous variable, and performance characteristics (including sensitivity, specificity and accuracy) are reported to complete this step. After an experiment runs on selected datasets, results should be compared to the expected target values defined in the “Qualification Data” file. Biomarker reproducibility in the clinical context is assessed using scans from patients that were imaged with the particular modality repeatedly and over an appropriately short period of time, without intervening therapy. The statistical approaches include standard analyses using intraclass correlation and Bland-Altman plots for the assessment of agreement between measurements. However, more detailed determinations are also of interest for individual biomarkers: For example, it may be useful to determine the magnitude of observed change in a biomarker that would support a conclusion of change in the true measurement for an individual patient. It may also be of interest to determine if two modalities measuring the same quantity can be used interchangeably. The diagnostic accuracy of biomarkers (that is, the accuracy in detecting and characterizing the disease) is assessed using methods suitable to the nature of the detection task, such as ROC, FROC, and LROC. In settings where the truth can be effectively considered as binary and the task is one of detection without reference to localization, the broad array of ROC methods will be appropriate. BBMSC Model View Controller SUC SUC SUC SUC SUC SUC SUC SUC SUC SUC SUC SUC 10 of 15 QI-Bench “Analyze” AAS Rev 0.4 Requirements Origin Since many imaging biomarkers in the volumetric analysis area produce measurements on a continuous scale, methods for estimating and comparing ROC curves from continuous data are needed. In settings where a binary truth is still possible but localization is important, methods from free-response ROC analysis are appropriate. Conduct pilot study(ies) of the analysis to establish capability of the class of tests that represent the biomarker using the training set (e.g., following Sargent et al., as set utility determinations restricted to demonstrating that in single studies the endpoint captures much of the treatment benefit at the individual patient level.) Demonstrating a high correlation at the patient level between the early endpoint and the ultimate clinical endpoint within a trial, randomized or not, is not sufficient to validate an endpoint. Such a correlation may be a result of prognostic factors that influence both endpoints, rather than a result of similar treatment effect on the two endpoints. Despite this caveat, a reasonably high patient level correlation (for example, >50%) would suggest the possible utility of the early endpoint and the value of subsequently assessing, by means of a larger analysis, the predictive ability of the early endpoint for the ultimate phase 3 endpoint for treatment effect at the trial level. For predictive markers, the Freedman approach involves estimating the treatment effect on the true endpoint, defined as s, and then assessing the proportion of treatment effect explained by the early endpoint. However, as noted by Freedman, this approach has statistical power limitations that will generally preclude conclusively demonstrating that a substantial proportion of the treatment benefit at the individual patient level is explained by the early endpoint. In addition, it has been recognized that the proportion explained is not indeed a true proportion, as it may exceed 100%, and that whilst it may be estimated within a single trial, that data from multiple trials are required to provide a robust estimate of the predictive endpoint. Additionally, it can have interpretability problems, also pointed out by Freedman. Buyse and Molenberghs also proposed an adjusted correlation method that overcomes some of these issues. For prognostic markers, the techniques for doing so are most easily described in the context of a continuous surrogate (e.g. change in nodule volume) and a continuous outcome. Linear mixed models with random slopes (or, more generally, random functions) and intercepts through time are built for both the surrogate marker and the endpoint. That is, the joint distribution of the surrogate marker and the endpoint are modeled using the same techniques as used for each variable individually. The degree to which the random slopes for the surrogate and the endpoint are correlated give a direct measure of how well changes in the surrogate correlate with changes in the endpoint. SUC BBMSC Model View Controller SUC SUC SUC SUC SUC SUC SUC SUC SUC SUC SUC SUC 11 of 15 QI-Bench “Analyze” AAS Rev 0.4 Requirements Origin The ability of the surrogate to extinguish the influence of potent risk factors, in a multivariate model, further strengthens its use as a surrogate marker. Conduct the pivotal analysis on the test set (extending the results to the trial level and establishing the achievable generalizability based on available data). Follow statisitical study designs as consistent with the claims and the type of biomarker along the lines described in the Basic Story Board for predictive vs. prognostic biomarkers. SUC Activity: Annotate biomarker db with results of group statistic SUC The class of tests serves as a basis for defining the measurement technology for a biomarker which may then be assessed as to its clinical utility. This assessment may be done in the context of an effort to qualify the biomarker for use in regulatory decision making in clinical trials, or it may be a comparable activity associated with individual patient management without explicitly following a qualification pathway. In either case, the hallmark of this step is the assessment of clinical utility on the basis of at least some capability to measure it. SUC Model View Controller EA SUC SUC 3.2. Performance Non-functional requirements, such as how fast a capability needs to run. Requirements Origin Comment / TI User interface display and update time needs to feel quick. AAS Implication is that can’t load whole ontologies but rather work incrementally. Design Guideline 3.3. Quality Control Support for internal and/or external quality control. Requirements Origin Comment / TI Design Guideline 3.4. “Super user” and Service Support Additional capabilities needed by privileged users. Requirements Origin Comment / TI Design Guideline Need a way to assess irregularites in QIBO … instances and to rectify them. BBMSC 12 of 15 QI-Bench “Analyze” AAS Requirements Rev 0.4 Origin Comment / TI Design Guideline 3.5. Upgrade / Transition Requirements to support legacy and/or prior versions. Requirements Origin Comment / TI Design Guideline Origin Comment / TI Design Guideline Need to devise a means for orderly update as QI-Bench Core concepts come and go 3.6. Security Applicable security compliance requirements. Requirements User certificate needs to distinguish between view-only, authorized to create Biomarker DB instances, authorized to edit existing instances, and to curate QIBO. BBMSC 13 of 15 QI-Bench “Analyze” AAS Rev 0.4 3.7. Requirements for Subsequent Iterations Defined in the SUC but not yet staged for implementation. Requirements Origin Estimate the confidence intervals on tumor measurements due to the selected independent variables, as measured by validated volume difference measures. Review statistics results to uncover promising hypotheses about the data. Typical methods include: Box plot, Histogram, Multi-vari chart, Run chart, Pareto chart, Scatter plot, Stem-and-leaf plot, Odds ratio, Chi-square, Median polish, or Venn diagrams. Reports are generated. SUC Guidelines of “good practice” to address the following issues are needed: (i) composition of the development and test data sets, (ii) data sampling schemes, (iii) final evaluation metrics such as accuracy as well as ROC and FROC metrics for algorithms that extend to detection and localization. With development/testing protocols in place, the user would be able to report the estimated accuracy and reproducibility of their algorithms on phantom data by specifying the protocol they have used. Furthermore, they would be able to demonstrate which algorithmic implementations produce the most robust and unbiased results (i.e., less dependent on the development/testing protocol). The framework we propose must be receptive to future modifications by adding new development/testing protocols based on up-to-date discoveries. Inter-reader variation indicates difference in training and/or proficiency of readers. Intra-reader differences indicate differences from difficulty of cases. SUC To show the clinical performance of an imaging test, the sponsor generally needs to provide performance data on a properly-sized validated set that represents a true patient population on which the test will be used. For most novel devices or imaging agents, this is the pivotal clinical study that will establish whether performance is adequate. (in addition ot other workflows) comparative analyses would be pursued that identify the relative advantages (or disadvantages as the case may be) of using this biomarker vs. another biomarker. Two specific examples that are currently relevant include spirometry vs. based lung densitometry, and use of diameter measurements on single axial slices as presently inculcated in RECIST. Ultimately, use of all putative imaging biomarkers are understood to be in relation to how it is done without benefit of the imaging biomarker and industry uptake for the biomarker requires an evaluation of relative performance against identified figures of merit. There are two approaches: either: SUC BBMSC Comment / TI Design Guideline SUC SUC SUC SUC SUC SUC SUC SUC SUC SUC 14 of 15 QI-Bench “Analyze” AAS Rev 0.4 Requirements Origin Follow workflow “Measure Correlation of Imaging Biomarkers with Clinical Endpoints” for each of the two biomarkers, the “new” and the “accepted” and assess the degree to which each (independently) correlates with the desired clinical endpoint. The comparison is framed “indirectly” in terms of how well each correlates. The one that correlates better is said to be superior. Alternatively, the two biomarkers may be compared directly by following workflow “Measure Correlation of Imaging Biomarkers with Clinical Endpoints” only for the new biomarker and replacing the target of the correlation to be the result of following workflow “Create Ground Truth Annotations and/or Manual Seed Points in Reference Data Set” in the Reference Data Sets according to the previously accepted biomarker. The comparison in this case is more direct, with the implication being that the biomarker which calls an event first is considered better. The caveat is that the accepted biomarker may not actually be correct; in fat, it may be that the reason that the new biomarker is proposed is to overcome some deficiency in the prior biomarker so a direct comparison may be inconclusive because the “truth” of the event called is not established nor is it clear what happens in those cases where one biomarker calls an event but the other does not. Which one is correct in this case. Indication of whether the candidate implementation complies with the Profile (which in turn specifies the targeted performance with respect to clinical context for use). any of these activities may be performed on behalf of one or more sponsors by trusted broker so as to protect individual identities and/or perform on sequestered data SUC Comment / TI Design Guideline SUC SUC SUC SUC 4. Deployment Model(s) Relevant and representative examples of deployment scenarios 5. Implementation Considerations and Recommendations for Technical Realization Identification of topics requiring elaboration in candidate solutions. This may be application-specific, deployment related, or non-functional This specification in the real world (e.g., relationships to existing infrastructure, other deployed services, dependencies, etc.) BBMSC 15 of 15