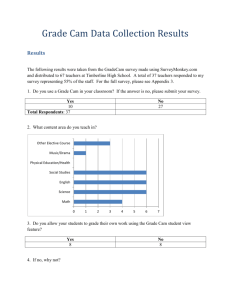
Media:Systems_Biology_Ch_5
advertisement

Chapter 5 - Signaling Networks L1 EGF P aPKC P aPKC Ca PKC P P Ca Kin ase Y891 PLC Ca Ca Ca Ca Ca Ca Ca Ca Ca GRK5 Y992 Y1045 Y1045 Y992 Y1086 Y1068 Y1068 Y1086 Y1173 Y1148 Y1148 Y1173 Fascin Integrin 1 PLC P BCL-2 S153 P RKIP CaM Raf1 14-3-3 KSR CaM MEK1/2 Tau CaM MLCK CaM DAPK Ca EF kinase CaM Ca CamKII CaM CnB NFAT P CamKII CaM CaM S217 S221 CaM PLC signaling module from http://visiscience.com/free_powerpoint_slides.php ERK1/2 P T202 P MKK4 P MEKK2 MKK7 P JNK1/2 S129 P NFAT GAP43 Y341 S338 CaM Phosphorylase CaM kinase Adducin MARCKS CamKIV CaM MAP2 Vinculin IP3 Ca Ca PKC DAG Y920 Y920 IP3 Dyna min-1 Syndecan-4 Y845 Y891 DAG EGF L2 CR2 Y845 PKC Ca P PKC Ca Ca L2 CR2 L1 Kin ase Ca CR1 CR1 CR1 P P P AP-1AP-1 Elk SRF P Ets Review: Chapters 1 & 2 Systems Biology Network Analysis Source: Palsoson, B. 2006. System Biology Network reconstruction Synthesis of in silico models Many states Different behaviors Multiple paths to accomplish same function Review: Chapter 3 Metabolic Networks Collection of pathways describing chemical modification Hierarchy – focus on level of individual reactions Pathways varied, but highly interconnected Source: Feigenson, G. 2006 BIOBM 331 Review: Chapter 4 Transcriptional Regulatory Networks “The on-off switches at the gene level” Determine genome expression state Networks are incompletely defined Approaches to network reconstruction in development Input Signals Regulating component Changed RNA and protein output Changed cell behavior and structures Adapted from http://genomicsgtl.energy.gov/science/generegulatorynetwork.shtml Chapter 5: Signaling Networks D + 1.5 E E-A E + 6C 2D + 3A A+B 2B + C Chapter 5: Signaling Networks Most Developed Reconstruction Metabolic Network Transcription Regulatory Network Signaling Network Least Developed What is signaling? Signaling – the transduction of a “signal” from outside of a cell to inside the cell. Signal – something that: the cell encounters in its environment causes a sequence of events results in a change in the cell What is Signaling? Example 1: Acetylcholine Ca Ca Ca Ca ACE Ca Ca Ca Ca Ca Ca Ca Ca Signal – something that: the cell encounters in its environment causes a sequence of events results in a change in the cell What is Signaling? Example 2: PLC EGF CR1 CR1 CR1 L2 CR2 •something the cell encounters in its environment Y845 Kin ase •causes a sequence of events L1 Y891 Y920 PLC L1 EGF EGF L2 CR2 Y845 Kin ase Signal Y891 Y920 Y992 Y1045 Y1045 Y992 Y1086 Y1068 Y1068 Y1086 Y1173 Y1148 Y1148 Y1173 PLC P •results in a change in the cell P P P P AP-1AP-1 Elk SRF P Ets from http://visiscience.com/free_powerpoint_slides.php Data Sources “High-throughput techniques are still in their infancy” Yeast two-hybrid assays Mass spec, isotope-coded affinity, target-assisted iterative screening Protein arrays Two-hybrid assay Image from http://www.scq.ubc.ca/?p=246 Fluorescence Resonance Energy Transfer (in development) FRET Image from http://mekentosj.com/science/fret/ What do we do with the Data? Integrate, Integrate, Integrate no single technique will provide enough information So far, work has been small scale Single receptor-ligand complex More and more data available, and progress is being made Why do we want to reconstruct biochemical networks Mathematically? Mathematics provides new insights. Reconstructing Signaling Networks Coarse level of detail (resolution) Finer Resolution E.g. ligand A and transcription factor B appear in a cell at about the same time E. g. ligand A causes a conformational change in transmembrane protein B, so protein B releases compound C, which activates protein D, which forms an association with protein E. This association causes protein F to dimerize, thus activating transcription factor G. Finest Resolution Mechanistic description of each individual reaction Reconstructing Signaling Networks Three general approaches so far: reconstructing highly connected nodes identifying signaling modules reconstructing pathways connecting signaling inputs and outputs D + 1.5 E E-A E + 6C 2D + 3A A+B 2B + C Building the Model: Pathway Example 1: Steroids S S S S S-Rec S S-Rec S S-Rec Trs Building the Model: Pathway Example 2: more PLC L1 CR1 CR1 CR1 L2 CR2 Kin ase Y845 Y891 Y920 PLC L1 EGF EGF L2 CR2 Y845 Kin ase EGF Y891 Y992 Y1045 Y1045 Y992 Y1086 Y1068 Y1068 Y1086 Y1173 Y1148 Y1148 Y1173 PKC DAG Y920 PLC P MKK4 P MEKK2 MKK7 P JNK1/2 S129 P P P P AP-1AP-1 Elk SRF P Ets from http://visiscience.com/free_powerpoint_slides.php Building the Model Pathways : more than spatial information Pathways consist of enzyme-catalyzed chemical transformations - this gives us stoichiometry Coupled chemical reactions imply Mass balance Thermodynamics Components – pathways – sectors -- whole cell functions Broad approaches, data, spatial and chemical relationships – feels good, right? Complications The signaling network is not static 1 Cell 1014 Cells There are lots of genes Images from http://en.wikipedia.org/wiki/Fertilisation http://www.erikandanna.com/OurNewBaby.htm http://folding.stanford.edu/education/GAH/gene.html More Complications Pathways may have multiple inputs and outputs, and components may be in multiple pathways. L1 L1 CR1CR1 E E L2 L2 GF CR2 CR2 GF Y891 Y891 DAG Dyna min-1 Syndecan-4 Vinculin Y845 Y845 Ki na se P aPKC P aPKC Ca PKCP Ca PKCP Ki na se Y920 Y920 PKC DAG Fascin Integrin 1 Adducin IP3 IP3 Y992Y1045Y1045Y992 PKCP Ca PLCP PLC Y1148Y1148 Y1173 Y1173 Ca Ca Ca Ca Ca P BCL-2 S153 Y1086 Y1068 Y1068 Y1086 RKIP GAP43 MARCKS Ca Ca Ca Ca Ca Ca Ca Ca GRK5 CaM CamKIVCaM DAPK CaM MAP2 CaM Tau CaM Ca MLCK CaM Y341 Raf1S338 KSR 14-3-3 Ca CaM Ca CamKIICaM EF kinaseCaM Phosphorylase CaM kinase CnB CaM P NFAT CamKIICaM NFAT MEK1/2 T202 ERK1/2 S217S221 P MKK4 P MKK7 MEKK2 JNK1/2 S129 P P P AP-1 AP-1 Elk P P SRF Ets from http://visiscience.com/free_powerpoint_slides.php P P Don’t be discouraged! Metabolic networks with more components have already been reconstructed Signaling networks have combinatorial features Closely interconnected network (coarse scale) Since transcription factors and signaling receptors associate, fewer are required Summary Signal transduction: signal to nucleus, or cytosol Formation of membrane complex (in most cases) Series of reactions Change in transcription Networks are combinatorial, dynamic Few pathways reconstructed Reconstruction involves multiple data types to create a mathematical model Quest ions? L1 EGF P aPKC P aPKC Ca PKC P P Ca Ca Kin ase Y891 PLC Ca Ca Ca Ca Ca Ca Ca Ca Ca GRK5 Y992 Y1045 Y1045 Y992 Y1086 Y1068 Y1068 Y1086 Y1173 Y1148 Y1148 Y1173 Fascin Integrin 1 PLC P BCL-2 S153 P RKIP CaM Raf1 14-3-3 KSR CaM MEK1/2 Tau CaM MLCK CaM DAPK Ca EF kinase CaM Ca CamKII CaM CnB NFAT P CamKII CaM CaM S217 S221 CaM PLC signaling module from http://visiscience.com/free_powerpoint_slides.php ERK1/2 P T202 P MKK4 P MEKK2 MKK7 P JNK1/2 S129 P NFAT GAP43 Y341 S338 CaM Phosphorylase CaM kinase Adducin MARCKS CamKIV CaM MAP2 Vinculin IP3 Ca Ca PKC DAG Y920 Y920 IP3 Dyna min-1 Syndecan-4 Y845 Y891 DAG EGF L2 CR2 Y845 PKC Ca P PKC Ca L2 CR2 L1 Kin ase Ca CR1 CR1 CR1 P P P AP-1AP-1 Elk SRF P Ets