Hints on Column Chromatography
advertisement

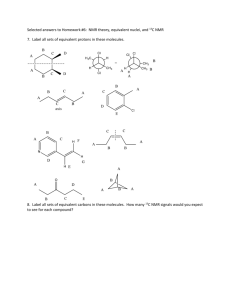
Lecture 3 NMR Spectroscopy: • Spin-spin Splitting in 1H NMR • Integration • Coupling Constants • 13C NMR • Sample Preparation for NMR Analysis Due: Lecture Problem 1 1H NMR Spectrum of Ethanol: Spin-Spin Splitting a b c a CH3CH2O H TMS a - triplet b - quartet c - singlet b c downfield ppm (d) upfield Spin-Spin Splitting a b c CH3CH2O H a - triplet b - quartet c - singlet General rules: • Neighboring, non-equivalent protons split each other’s signals • Equivalent protons do not split each other’s signals • Use the n + 1 rule to predict the splitting pattern of a proton’s signal n + 1 rule The signal of a proton with n equivalent neighboring protons is split into a multiplet of n + 1 peaks. In ethanol, a neighbors b; they split each other’s peaks. Note that b neighbors c and no splitting occurs between the two; b is only affected by a. In general, protons that reside on heteroatoms (O, N) do not get involved with spin-spin splitting with neighboring protons. Thus, c appears as a singlet. Spin-spin Splitting Spin-Spin Splitting Determine the splitting patterns for the signals in the 1H NMR spectra of the following compounds. OH O H Cl NH2 CH3 NH2 Complex Spin-Spin Splitting Consider the 1H NMR spectrum of a substituted alkene: Spin-Spin Splitting Spin-Spin Splitting Determine the splitting patterns for the signals in the 1H NMR spectra of the following compounds. O O O H H3C Cl H Integration • Area underneath signal; NMR machine will give integrals • First, gives the relative ratio of different types of protons in compound • Second, allows determination of actual ratio of different types of protons 1. Measure the length of the integral with a ruler 2. Establish a relative ratio of protons (divide each length by the lowest number) Coupling Constants (J) Protons that split each other’s peaks will have the same coupling constant or J value. 1H NMR Spectrum of a Taxol Derivative QuickTime™ and a TIFF (LZW) decompressor are needed to see this picture. Taken from Erkan Baloglu’s Masters Thesis Nuclear Magnetic Resonance Information Gained: • Different chemical environments of nuclei being analyzed (1H nuclei): chemical shift • The number of different types of H’s: number of signals in spectrum • The numbers of protons with the same chemical environment: integration • The number of protons are bonded to the same carbon: integration • The number of protons that are adjacent to one another: splitting patterns • The exact protons that are adjacent to one another: coupling constants 13C NMR Spectroscopy Information Gained: • Different chemical environments of carbons in molecule: chemical shift • The number different types of C’s: number of signals in spectrum Differences from 1H NMR: • No splitting of signals (proton-decoupled); thus, only singlets • No integration • ppm scale ranges from 0 to 220 ppm 13C Carbonyl Carbons NMR Chemical Shifts Unsaturated Carbons 200 C-X Saturated Carbons 0 100 downfield TMS upfield ppm Like with 1H NMR, the more shielded the carbon nuclei, the more upfield its signal will appear and vice versa. 13C NMR Spectrum of Chlorohexane 13C NMR Correlation Chart NMR Sample Preparation & CDCl3 Sample Prep: Dissolve ~32 mg of sample in CDCl3 in an NMR tube. Why use CDCl3? Deuterated solvents are necessary in NMR because deuterium is NOT NMR active and will not interfere with your sample’s spectrum. CDCl3 is 98-99% pure with a trace amount of CHCl3. You will see a small solvent peak at ~7.26 ppm due to CHCl3 (1H NMR); see a triplet at 77 ppm 13C NMR. This peak serves as a reference peak; DO NOT count it as one of your sample’s signals!