File
advertisement

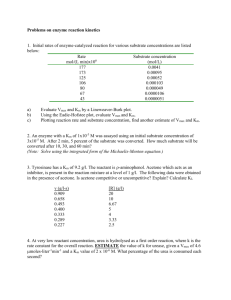
* Chapter 3 * * Likelihood that a product will form when a substrate molecule enters the active site * Sometimes, with low enzyme affinity, a substrate will enter an active site but instead of making a product, substrate will leave the active site before a reaction takes place and a product forms * * The rate at which an enzyme can convert a substrate molecule into product per second * Relies heavily on amount of collisions between enzyme and substrate molecule * Maximum number of chemical conversions of substrate molecules per second that a single catalytic site will execute for a given enzyme concentration * “Kcat” in formulas * * Vmax theoretical maximum rate of an enzyme ; tells us how well an enzyme performs * At Vmax ALL enzyme molecules are bound to substrate molecule * Enzyme is SATURATED with substrate * How to Measure Vmax: * Reaction rate is measured at different substrate concentrations while keeping enzyme concentration constant * As substrate concentration INCREASES reaction rate rises until it reaches maximum rate (Vmax) * Vmax is where your graph levels off… * Find the y-value for the plateau…this is Vmax * * Plotted against substrate concentrations * Produces ASYMPTONIC curve * In practice: * Curve never truly flattens out * In theory: * Curve flattens out when there is an infinite substrate concentration * Impossible to read a value of Vmax off a graph…so we must use another graph…. * * Create a double reciprocal plot (aka Lineweaver-Burke plot) * Plot inverse of substrate concentration on x-axis (1/[S]) * Plot inverse of velocity on y-axis (1/velocity) * Benefits: * 1/infinity substrate concentration = ZERO * Zero can be plotted on your graph * Vmax can be accurately found off of graph * Resulting line is a straight line (not curve) * Accurately determine Vmax * Accurately determine Km (Michaelis-Menton Constant) * Steps for Finding Vmax from a Double-Reciprocal Plot * Find 1/Vmax * Point where the line crosses/intersects the y-axis * Point of intersection on y-axis is where 1/[S] is equal to zero (when [S] is infinity) * Once 1/Vmax is determined, Vmax can be calculated * Km (Michaelis Menten Constant) can also be determined from the double reciprocal plot * * Km (abbreviation) * Measure of the affinity of an enzyme • Higher affinity= lower Michaelis-Menton constant = for its substrate reaction will occur much more quickly (towards its * The substrate concentration at which an enzyme works at HALF its maximum Vmax) rate (1/2 Vmax) • Lower affinity = higher * At this point, HALF the active sites of Micahelis-Menton constant = the enzymes are occupied by the reaction will occur much substrate more slowly * Higher affinity of the enzyme for the substrate lower concentration is needed for Km • Low Km = High affinity of enzyme for substrate = more product • High Km = Low affinity of enzyme for substrate = less product * * Use double reciprocal plot * -1/Km is the point where the line for the graph intersects x-axis * Use the value of -1/Km to calculate Km * Value of Km can vary depending on many factors * Identity of substrate * Temperature * pH * Presence of particular ions * Overall ion concentration * Presence of poisons * Presence of pollutants * Presence of inhibitors * Vmax * * Info about maximum rate of reaction that is possible * Km * Measures affinity of the enzyme for the substrate * Higher affinity = more likely product will be formed when substrate enters active site * High affinity produces LOW Km value * Low affinity produces HIGH Km value… * What would give you a HIGH Km value (low affinity for substrate to go to active site)? * INHIBITORS!!!! * * The smaller the 1/Km value is larger the Km value is * The larger the 1/Km value is smaller Km value is * The smaller the 1/Vmax value is larger the Vmax value is * The larger the 1/Vmax value is smaller Vmax value is Large Vmax means you need to add lots a substrate to make reaction happen faster Small Vmax means only a little substrate is need to make reaction happen fast Large Km value LOW enzyme affinity (some reason, the substrates do NOT want to bind to active site) Low Km Value HIGH enzyme affinity (substrates immediately bind to active sit of enzymes…only a small substrate concentration is needed to saturate your enzymes’ active sites) * Steps: * Create table recording “v” (velocity) vs. substrate concentration [S] * Plot rate of reaction “v” (velocity) vs. substrate concentration [S] * V y-axis * [S] x-axis * Can’t tell Vmax from graph…. * Revisit table and now fill in additional columns, adding 1/v and 1/[S] by doing simple calculations * Now, make a new graph * Plot 1/v on y-axis and 1/[S] on x-Axis * Plot values from table on this new graph * Draw straight line through points….ALL the way through y-axis and onto negative side of x-axis * Your y-intercept is the 1/Vmax when the 1/[S] is 0 (which, do the math, would equal INFINITY!!!! Endless substrate, yay!) * Your x-intercept is the 1/km * Plug in these values into the above equations, solve for Km and Vmax, now you have determined Vmax for your original graph!!! [S]/ 1/[S]/ v/ 1/v / arbitrary unit arbitrary unit arbitrary unit arbitrary unit * *