What effects do salts have on biopolymers?
advertisement
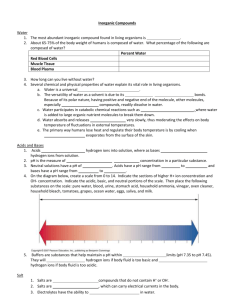
What effects do salts have on biopolymers? . Maxim V. Fedorov1, Ingrid Socorro1, Stephan Schumm2 and Jonathan M. Goodman1 1 Unilever Centre for Molecular Science Informatics, Department of Chemistry, University of Cambridge, Cambridge, UK 2 Unilever R&D Vlaardingen, Vlaardingen, The Netherlands • Research: Biopolymers interactions with salts in water. • Systems: Water with salt and: – Oligopeptides (3 - 21 amino acids) – Bee toxin (mellitin; 27 amino acids) • Methods: Fully atomistic Molecular Dynamics simulations What effect do salts have on biopolymers? Some of important processes are: - biopolymer solubility - biopolymer denaturation temperatures - enzyme activity - biopolymer swelling - growth rates of bacteria - stability of protein macroaggregates Energetic optimization of mutual hydrogen bonded networks The water hydrogen bonded network links secondary structures within the protein Water and cosolutes as ‘lubricants’ for protein folding. Schematic potential energy funnel for the folding of proteins without water: many barriers to the preferred minimum energy structure on the folding pathway There are numerous local minima that might trap the protein in an inactive three-dimensional molecular conformation Fully hydrated protein: the potential energy landscape is smoother Salt effects on biopolymer shapes D. Puett et al., JCP, 1967; H. Saito et al., Biopolymers, 1978; K. Zero & B.R. Ware, JCP, 1984; Poly-L-Lysine, bulk water solution Poly-L-Lysine, NaCl solution Assembly of two oligopeptides in water C. Muhle-Goll, et al., Biochemistry, 1994; P. Aymard, D. Durand & T. Nicolai, Int. J. Biol. Macromol.,1996 Self-assembly to very stable amyloids M. V. Fedorov et al Phys. A, 2006 • Poly-L-Lysine (PLL), a-Helix • Water solution (2300 water molecules) • Water and salt (2300 water and a few dozen NaCl molecules) Results: No Ions R A N D O M C O I L Comparison of two systems PLL: no ions PLL: 0.50 M NaCl Ramachandran density maps 0.50 M NaCl Na+ H E L I X Cl- Three major effects of ions • Electrostatic screening (Debye-Hückel effect) • Specific interactions by ion-pair formation (Electroselectivity effect) • Salts affect water structure, which may change the hydrophobic interactions (Hofmeister effect) Hofmeister Effect In 1888 Hofmeister observed that protein solubilities are influenced by the concentration and type of salts present. Solubilities tend to follow the general order: SO42- < F- < Cl- < Br- < I- < SCN- < ClO4precipitate, stabilize solubilize, destabilize Mg2+ < Na+ < K+ < Li+ Other phenomena to follow the Hofmeister series: - water activity - self-diffusion coefficient of water - viscosity of salt solutions - surface tension - lipid solubility of monoanions - polymer cloud points - polymer swelling - protein solubility - protein denaturation temperatures - degree of protein aggregation - coacervate behaviour - critical micellar concentration - enzyme activity - growth rates of bacteria Electroselectivity (direct binding) effect In 1990-1992 Goto and coworkers observed that conformation properties of some polypeptide and proteins solubilities are influenced by the concentration and type of 1:1 salts present following the electroselectivity series: electroselectivity I- < Br- < Cl- < F- precipitate, stabilize solubilize, destabilize Hofmeister F- < Cl- < Br- < IInverse order with compare to the Hofmeister series for monovalent anions. Verification • If the Debye-Huckel screening is important, the effect of various ions will be determined only by the ionic strength of solution • If the electroselectivity is important, the effect of different ions should follow the electroselectivity series of the salts • The importance of the Hofmeister effect can be determined by comparing the different ions with the Hofmeister series System • • • • • • • • • • • Poly-L-Alanine 3 (PAA), a-Helix (-57,-47). ~1200 water molecules (TIP5P-E) Li+, Na+, K+, Cs+ cations F-, Cl-, Br-, I- anions. OPLS/AA force-field Periodic Boundary Conditions Particle Mesh/Ewald electrostatics BOX: 37 Å x 37 Å x 37 Å Berendsen thermostat/barostat GROMACS 3.3 Equilibration run: 27 ns, production run: 27 ns. + - How long shall we simulate? Sampled part V of the available volume Vmax (volume of the box without the excluded volume of the tripeptide) visited be any of chlorine ions as a function of simulation time. This demonstrates equilibration time >> 1 ns as required for chlorines to visit any part of the box. Molecular Surfaces • Dotted line: Solvent Accessible Surface (SAS) • Solid line: molecular surface (MS) • Shaded grey area: van der Waals surface Water accessible area Compact conformations Water accessible area NaCl NaBr NaI NaF Ratio of compact conformations WHY??? Electroselectivity (direct binding) effect electroselectivity I- < Br- < Cl- < F- precipitate, stabilize solubilize, destabilize Hofmeister F- < Cl- < Br- < IInverse order with compare to the Hofmeister series for monovalent anions. Possible ways of interactions: • Direct contacts: + - • Shell – Shell contacts: -- + • Site – water – Site contacts: + - + + - Direct Contact: fluorine anions Shell-shell interactions. Macroscopic Coulomb’s law doesn’t work on the molecular level. • Macro q U macro ( r ) 0r Micro q U micro (r ) F (r ) 0 r F(r) – dielectric permittivity (screening factor). F (r ) 1, r Visualization of the (a) electrostatic potential, (b) screening factor and (c) ion-hydrogen (- - -) and ion-oxygen ( --- ) RDF, created by a single charge anion/ cation (of radii a =1 A ) as functions of the distance from the surface of the sphere, R-a, which mimics the ion:. MVF and A. A. Kornyshev, Mol. Phys., 2007, to be issued soon Potential of Mean Force Activation Energy Structure & Dynamics Peptide-water and ion-water PMFs EIW Peptide –ion PMFs N-terminus C-terminus Peptide –ion PMFs Side chain groups Backbone groups EPI Preferential Interaction Cosolutes will change the chemical potential of a protein in a cosolvent solution compared to a pure solvent due to preferential interaction with or exclusion from the protein interface. Gtr 0 Gtr 0 transfer free energy: The protein prefers to be surrounded by cosolvent molecules Protein solvent Gtransfer cosolvent protein protein water cosolvent The transfer from pure solvent to the cosolvent solution in unfavourable. The protein prefers to be surrounded by solvent molecules. Protein Specific interactions of ions with polypeptide charges (local effects) Preferential solvent / salt interactions (bulk effects) Conclusions • Salt effects on biopolymer solutions can be reproduced “in silico” using the fully atomistic Molecular Dynamics simulations. • Generally, ions contact biopolymers via an intermediate water shell. • We can distinguish between the Debye-Hückel, Electroselectivity and Hofmeister effects for salts Financial Support • Unilever R&D Vlaardingen Acknowledgements Robert Glen, Dmitry Nerukh ( Unilever Centre for Molecular Science Informatics, University of Cambridge, UK); Ruth Lynden-Bell (University of Belfast & University of Cambridge, UK); Alexei A Kornyshev (Imperial College London, UK); Gennady N Chuev (Institute of Experimental and Experimental Biophysics of RAS, Pushchino Biological Centre, Russia & University of Edinburgh, UK).