Chapter 6
advertisement

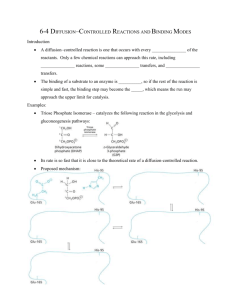
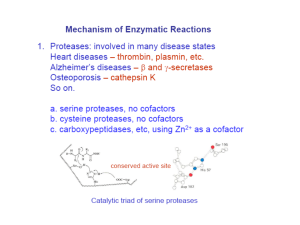
Enzyme Rate Enhancement How do enzymes work to catalyze reactions? Enzyme Classes by Reaction Type Oxidation-Reduction Reaction: Oxidoreductase Functional Group Transfer: Transferase Cutting with Water: Hydrolysis Group Elimination with Double Bond Formation: Lyase Compound Conversion to an Isomeric Form: Isomerase Triose phosphate isomerase Bond Formation with ATP Hydrolysis: Ligase DNA Ligase -- nucleotide base coupling with loss of PPi Chymotrypsin Catalyzes Peptide Hydrolysis Enzyme specificity in terms of: Site of hydrolysis and Substrate reactivity How can a reaction be pushed in the forward direction? Reactions that are Spontaneous in the Forward and Reverse Direction How do enzymes affect the energy diagram of a reaction? Enzymes Decrease the Activation Energy How can enzymes lower the transition state? Is external energy required? Mechanisms of Enzyme Catalysis Acid-base, covalent, metal ion, orientation and proximity Ketone-Enol Conversion What mechanism of enzyme catalysis is operative here? Amino Acid Residues that Function in Acid-Base Catalysis Conventions for Depicting Reaction Mechanisms Curved arrow indicates electron rearrangement during a reaction Covalent Catalysis A formal positive charge favors electron flow to the nitrogen group Covalent Catalysis Amino Acids that can Participate in Covalent Catalysis Metal Ion Catalysis Components that Facilitate Enzyme Catalysis What is the cofactor in ATP hydrolysis? What is the co-substrate in an oxidation-reduction reaction? Enzymes Decrease the Activation Energy Mechanisms of Enzyme Catalysis • Acid-Base Catalysis • Covalent Catalysis • Metal Ion Catalysis • Orientation/Proximity Effects • Preferential Transition- State Binding Proximity and Orientation Effects Facilitate Catalysis Chymotrypsin Specificity Cleaves peptides on the C-terminus side of hydrophobic residues (e.g. Phe, Tyr and Try) Active Site Mapping via Irreversible Inhibitors Diisopropylphosphofluoridate (DIPF) inhibits chymotrypsin by modifying 1 of 28 serine residues Active Site Mapping via Irreversible Inhibitors Covalent Catalysis for Chymotrypsin: a Two Step Process • Enzyme acylation with leaving group departure • Enzyme deacylation What is the leaving group with Chymotrypsin? Chymotrypsin Catalysis Proceeds via a Two-Step Mechanism Chromogenic substrate for kinetic studies Why is this compound not an ideal substrate mimic? Chymotrypsin Catalytic Triad • Catalytic triad serves as the site of catalysis • Aspartate and histidine contribute serine’s basicity • Serine serves as a nucleophile in covalent catalysis What type of catalysis occurs? Mechanism of Peptide Hydrolysis in Chymotrypsin Substrate binding via nucleophilic attack Mechanism of Peptide Hydrolysis in Chymotrypsin Polypeptide original C-side serves as leaving group Mechanism of Peptide Hydrolysis in Chymotrypsin Water attacks original Nside of polypeptide Mechanism of Peptide Hydrolysis in Chymotrypsin Polypeptide original N-side serves as leaving group and enzyme is regenerated Chymotrypsin Hydrolysis Tetrahedral-Intermediate Stabilization in Chymotrypsin H-bonds ideally positioned in the oxyanion hole stabilize the sp3 transition state Chymotrypsin Specificity Pocket Large structural pocket lined with hydrophobic amino acids favors bulky hydrophobic residues Serine Proteases Differ in Little Except Their Specificity Pockets Chymotrypsin Trypsin Elastase Substrate Specificity Observed with each Proteolytic Enzyme • Papain cleave peptides non-selectively • Trypsin cleaves carboxyl side of bulky + charged Rgroups • Chymotrypsin cleaves carboxyl side of bulky aromatic R-groups Thrombin Divergent Evolution Percent Sequence Identity among Three Serine Proteases Bovine trypsin Bovine chymotrypsinogen Porcine elatase Common ancestor with retention of overall structure and catalytic mechanism 100% 53% 48% Convergent Evolution Bovine versus bacterial serine protease No sequence or structural similarity but the same catalytic triad and oxyanion hole in the active site Enzyme-Substrate Binding Critical for Catalysis Lock and Key Model Enzyme Active Site • 3-D cleft or crevice • Small part of enzyme • Unique micro-environment • Substrate binding by weak forces Induced Fit Model Substrate-Induced Enzyme Conformational Change Substrate (-) Substrate (+) Inhibition by Transition State Analogs Pyrrolidine the natural substrate binds 160 less tightly than pyrrole a transition state analog. What is the favored enzyme binding geometry? Rate of Enzyme Catalysis Explain why enzyme activity increases with temperature and then precipitously drops off RNAas A Digestive Enzyme Cleaving Mechanism Why does ribonuclease catalyzes the hydrolysis of RNA but not DNA Conversion of Adenosine to Inosine What does the much greater binding affinity of 1,6dihydropurine ribonucleoside than the substrate indicate about the enzyme mechanism? Chapter 6 Problems: 1, 3, 7, 9, 11, 15, 20, 23, 25 and 37