Genetics
advertisement
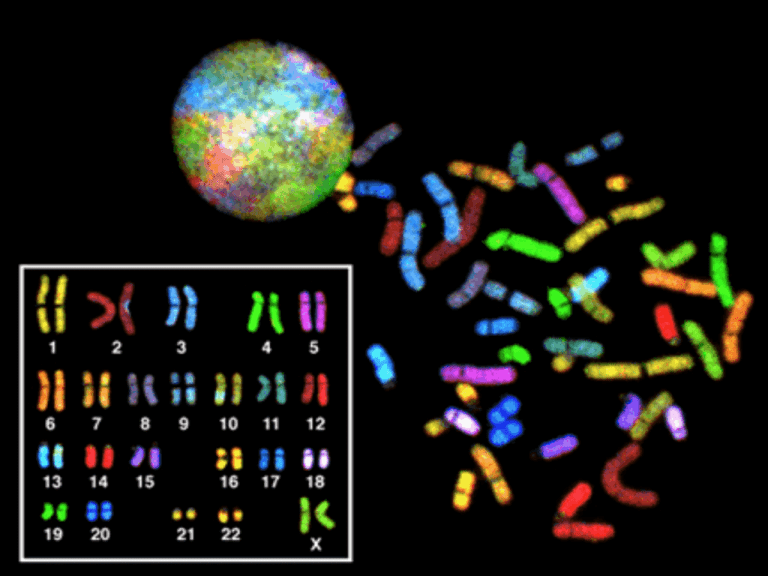
GENETIC DISORDERS DISEASES •GENETIC •ENVIRONMENTAL •BOTH • CONGENITAL • HEREDITARY • FAMILAL MUTATIONS • PERMANENT change in DNA • GENE MUTATION: (may, and often, result in a single base error) • CHROMOSOME MUTATION: (visible chromosome change)part of chromosome • Translocation • inversions • GENOME MUTATION: (whole chromosome) Base pair triplet gene chromosome segment whole chromosome genome COMPLEX MULTIGENIC DISORDER • Interaction btw varient forms of genes and environmental factors • Gene variation---polymorphism----multigenic or polygenic( atherosclerosis, diabetes, hypertension, ht &WT • CONSTITUTIONAL germ cells inherited disorder—in all cells • SOMATIC → cancer ↘congenital malformation in specific cells • PENETRATION GENE MUTATION • DELETION OF A SINGLE BASE • SUBSTITUTION OF A SINGLE BASE • POINT MUTATION---WITHIN CODING SEQUENCE--single base/different base--• Results with replacement of one amino acid with another 1. Missence mutation conservative nonconservative 2.Nonsense mutation---AA change—stop codon—Beta thal POINT MUTATION • MUTTIONS in NON-coding sequences defective transcription, regulation, apop. • DELETIONS/INSERTIONS 1.Multiple of three • 2.Premature stop codon • 3.“frameshift” mutation, involvement is NOT a multiple of 3 • Tri-nucleotide REPEATS, e.g., CGG repeats many times in fragile X syndrome, CAG in others SEQUENCE AND COPY NUMBER VARIATIONS (POLYMORPHISMS) • any two individuals share greater • than 99.5% of their DNA sequences • diversity of humans is encoded in less than 0.5% of our • , this 0.5% represents about 15 million base • pairs. • two most common forms of DNA variations Two most common forms of DNA variations (polymorphisms) in the human genome are 1.Singlenucleotide polymorphisms (SNPs) SNPs represent variation at single isolated nucleotide 2.Copy number variations (CNVs). • • • positions and are almost always biallelic (i.e., one of only • two choices exist at a given site within the population, • such as A or T). Much effort has been devoted to making • SNP maps of the human genome. These efforts have • identified over 6 million SNPs in the human population, • CNVs are a recently identified form of genetic variation • consisting of different numbers of large contiguous • stretches of DNA from 1000 base pairs to millions of • base pairs. In some instances these loci are, like SNPs, • biallelic and simply duplicated or deleted in a subset of Epigenetic Changes • Epigenetic changes are those involving modulation of gene or protein expression in the absence of alterations in DNA sequence (i.e., mutation) • Epigenetic regulation is of critical importance during development, as well as in homeostasis of fully developed tissues. • alterations in the methylation of cytosine residues at gene promoters—heavily methylated promoters become inaccessible to RNA polymerase, leading to transcriptional • silencing. Promoter methylation and silencing of tumor suppressor genes leads to unchecked cell growth –cancer • Another major player in epigenetic are • histone proteins, which are components of structures called nucleosomes, around which DNA is coiled. • Histone proteins undergo a variety of reversible modifications (e.g., methylation, • acetylation) that affect secondary and tertiary DNA structure, and hence gene • Alterations in Non-Coding RNAs • —so-called “non-coding RNAs (ncRNAs)”— play important regulatory functions. • Although many distinct families of ncRNAs exist • two imortant are microRNAs (miRNAs), and long non-coding RNAs • The miRNAs, unlike messenger RNAs, do • not encode proteins but instead inhibit the translation of target mRNAs into their corresponding proteins. Posttranscriptional • silencing of gene expression by miRNA is preserved in all living forms from plants to humans and is therefore a fundamental mechanism of gene regulation • In addition to alterations in DNA sequence, coding genes also can undergo structural variations, such as copy number changes (amplifications or deletions), or translocations, • resulting in aberrant gain or loss of protein function. with mutations, Philadelphia chromosome— translocation t(9;22) between the BCR and ABL genes in • chronic myelogenous leukemia GENE MUTATIONS • • • • • INTERFERE with protein synthesis SUPPRESS transcription, DNARNA PRODUCE abnormal mRNA DEFECTS carried over into TRANSLATION ABNORMAL proteins WITHOUT impairing syntheses GENETIC DISORDERS • SINGLE gene mutations, following classical MENDELIAN inheritance patterns the most • MULTIFACTORIAL inheritance • CHROMOSOMAL disorders • NON-MENDELIAN disorders MENDELIAN inheritance patterns • AUTOSOMAL DOMINANT • AUTOSOMAL RECESSIVE • SEX-LINKED (recessive), involving “X” chromosome • • Suspected sex chromosome abnormality (e.g., Turner syndrome) • • Suspected fragile X syndrome • • Infertility (to rule out sex chromosome abnormality) • • Multiple spontaneous abortions (to rule out the parents as carriers of balanced translocation; AUTOSOMAL DOMINANT • Disease is in HETEROZYGOTES • NEITHER parent may have the disease (NEW mut.) • REDUCED PENETRANCE (environment?, other genes?) • VARIABLE EXPRESSIVITY (environment?, other genes?) • May have a DELAYED ONSET • Usually result in a REDUCED PRODUCTION or INACTIVE protein AUTOSOMAL DOMINANT • • • • • • • HUNTINGTON DISEASE NEUROFIBROMATOSIS MYOTONIC DYSTROPHY TUBEROUS SCLEROSIS POLYCYSTIC KIDNEY HEREDITARY SPHEROCYTOSIS VON WILLEBRAND DISEASE • • • • • • MARFAN SYNDROME EHLERS-DANLOS SYNDROMES (some) OSTEOGENESIS IMPERFECTA ACHONDROPLASIA FAMILIAL HYPERCHOLESTEROLEMIA ACUTE INTERMITTENT PORPHYRIA AUTOSOMAL DOMINANT PEDIGREE 1) BOTH SEXES INVOLVED 2) GENERATIONS NOT SKIPPED AUTOSOMAL RECESSIVE • Disease is in HOMOZYGOTES • More UNIFORM expression than AD • Often COMPLETE PENETRANCE • Onset usually EARLY in life • NEW mutations rarely detected clinically • Proteins show LOSS of FUNCTION and compensated in heterozygote form • Include ALL inborn errors of metabolism • MUCH more common that autosomal dominant AUTOSOMAL RECESSIVE • • • • • • • • • CF PKU GALACTOSEMIA HOMOCYSTINURIA LYSOSOMAL STORAGE Α-1 ANTITRYPSIN WILSON DISEASE HEMOCHROMATOSIS GLYCOGEN STORAGE DISEASES Hgb S THALASSEMIAS CONG. ADRENAL HYPERPLASIA EHLERS-DANLOS (some) ALKAPTONURIA NEUROGENIC MUSC. ATROPHIES FRIEDREICH ATAXIA SPINAL MUSCULAR ATROPHY AUTOSOMAL RECESSIVE PEDIGREE 1) BOTH SEXES INVOLVED 2) GENERATIONS SKIPPED SEX (“X”) LINKED • • • • MALES ONLY HIS SONS are OK, right? ALL his DAUGHTERS are CARRIERS The “Y” chromosome is NOT homologous to the “X”, i.e., the classic concept of dominant/recessive has no meaning here • HETEROZYGOUS FEMALES have no phenotypic expression (carriers)….usually, this means autosomal “recessive”, right? SEX (“X”) LINKED • • • • • • • • DUCHENNE MUSCULAR DYSTROPHY HEMOPHILIA , A and B G6PD DEFICIENCY AGAMMAGLOBULINEMIA WISKOTT-ALDRICH SYNDROME DIABETES INSIPIDUS LESCH-NYHAN SYNDROME FRAGILE-X SYNDROME SEX LINKED PEDIGREE 1) MALES ONLY, sons of affected males are OK 2) GENERATION SKIPPING DOESN’T MATTER SINGLE GENE DISORDERS • ENZYME DEFECT (Most of them, e.g., PKU) – Accumulation of substrate – Lack of product – Failure to inactivate a protein which causes damage • RECEPTOR/TRANSPORT PROTEIN DEFECT (Familial Hypercholesterolemia) • STRUCTURAL PROTEIN DEFECT (Marfan, Ehl-Dan) – Structure – Function – Quantity • ENZYME DEFECT WHICH INCREASES DRUG SUSCEPTIBILITY: G6PDPrimaquine STRUCTURAL PROTEIN DEFECTS • Marfan Syndrome – Fibrillin-1 defect (not -2 or -3) – Tall, dislocated lens, aortic arch aneurysms, etc. – Abraham Lincoln?, Osama bin-Laden? • Ehlers-Danlos Syndromes (AD, AR) – Multiple (6?) different types – Classical, Hypermob., Vasc., KyphoSc., ArthChal., Derm – Various collagen defects – Hyperelastic skin, hyperextensible joints RECEPTOR PROTEIN DEFECTS • FAMILIAL HYPERCHOLESTEROLEMIA – LDL RECEPTOR defect – Cholesterol TRANSPORT across liver cell impaired – ergo, CHOLESTEROL BUILDUP IN BLOOD • “Scavenger System” for CHOL kicks in, i.e., MACROPHAGES • YOU NOW KNOW THE REST OF THE STORY • YOU NOW KNOW WHY MACROPHAGES are “FOAMY” ENZYME DEFICIENCIES • BY FAR, THE LARGEST KNOWN CATEGORY – SUBSTRATE BUILDUP – PRODUCT LACK – SUBSTRATE could be HARMFUL • LYSOSOMAL STORAGE DISEASES comprise MOST of them LYSOSOMAL STORAGE DISEASES • • • • • • GLYCOGEN STORAGE DISEASES SPHINGOLIPIDOSES (Gangliosides) SULFATIDOSES MUCOPOLYSACCHARIDOSES MUCOLIPIDOSES OTHER – Fucosidosis, Mannosidosis, Aspartylglycosaminuria – WOLMAN, Acid phosphate deficiency GLYCOGEN STORAGE DISEASES • MANY TYPES (at least 13) • Type 2 Pompe (acid-α-glucosidase) , von Gierke (Glu-6P-ase), McArdle (phosphorylase), most studied and discussed, and referred to • Storage sites: Liver, Striated Muscle (Skel + Ht) SPHINGOLIPIDOSES • MANY types, Tay-Sachs most often referred to – – – – – GANGLIOSIDES are ACCUMULATED Ashkenazi Jews (1/30 are carriers) CNS neurons a site of accumulation CHERRY RED spot in Macula Usually fatal by age 4 SULFATIDOSES • MANY types, but the metachromatic leukodystrophies (CNS), Krabbe, Fabry, Gaucher, and Niemann-Pick (A and B) are most commonly referred to • SULFATIDES, CEREBROSIDES, SPHINGOMYELIN are the accumulations • • • • • • NIEMANN-PICK TYPES A, B, C SPHINGOMYELIN BUILDUP Sphingomyelinase (ASM), is the missing enzyme MASSIVE SPLENOMEGALY ALSO in ASHKANAZI JEWS OFTEN FATAL in EARLY LIFE, CNS, ORGANOMEGALY GAUCHER DISEASE • GLUCOCEREBROSIDE BUILDUP • 99% are type I, NO CNS involvement • ALL MACROPHAGES, liv, spl, nodes, marrow MUCOPOLYSACCHARIDOSES • HURLER/HUNTER, for I and II, respectively, 14 types • DERMATAN sulfate, HEPARAN sulfate buildup, respectively – coarse facial features – clouding of the cornea – joint stiffness – mental retardation – URINARY EXCRETION of SULFATES COMMON OTHER LYSOSOMAL STORAGE DIS. • • • • • FUCOSIDOSIS MANNOSIDOSIS ASPARTYLGLYCOSAMINURIA WOLMAN (CHOL., TRIGLYCERIDES) ACID PHOSPHATE DEFICIENCY (PHOS. ESTERS) ALCAPTONURIA • • • • NOT a LYSOSOMAL ENZYME DISEASE FIRST ONE TO BE DESCRIBED HOMOGENTISIC ACID HOMOGENTISIC ACID OXIDASE –BLACK URINE –BLACK NAILS (OCHRONOSIS), SKIN –BLACK JOINT CARTILAGE (SEVERE ARTHRITIS) NEUROFIBROMATOSIS • 1 and 2 – 1-von Recklinghausen – 2- “acoustic” neurofibromatosis • 1 – Neurofibromas, café-au-lait, Lisch nodules NEUROFIBROMATOSIS • 1 and 2 • 1-von Recklinghausen • 2- “acoustic” neurofibromatosis • 2 – Bilateral acoustic neuromas and multiple meningiomas MULTIFACTORIAL INHERITANCE • Multi-”FACTORIAL”, not just multi-GENIC • “SOIL” theory • Common phenotypic expressions governed by “multifactorial” inheritance – Hair color – Eye color – Skin color – Height – Intelligence – Diabetes, type II FEATURES of multifactorial inheritance • • • • Expression determined by NUMBER of genes Overall 5% chance of 1st degree relatives having it Identical twins >>>5%, but WAY less than 100% This 5% is increased if more children have it • Expression of CONTINUOUS traits (e.g., height) vs. DISCONTINUOUS traits (e.g., diabetes) “MULTIFACTORIAL” DISORDERS • • • • • • • • Cleft lip, palate Congenital heart disease Coronary heart disease Hypertension Gout Diabetes Pyloric stenosis MANY, MANY, MANY, MANY MORE….. • • • • KARYOTYPING Defined as the study of CHROMOSOMES 46 = (22x2) + X + Y Conventional notation is “46,XY” or “46,XX” G(iemsa)-banding, 500 bands per haploid recognizable • Short (“p”-etit) arm = p, other (long) arm = q More KARYOTYPING info • A,B,C,D,E,F,G depends on chromosome length – A longest – G shortest • Groups within these letters depend on the p/q ratio • ARMREGIONBANDSub-BAND, numbering from the centromere progressing distad F.I.S.H. (gene “probes”) greatly enhances G-banding • Fluorescent InSitu Hybridization • Uses fluorescent labelled DNA fragments, ~10,000 base pairs, to bind (or not bind) to its complement FISH • SUBTLE MICRODELETIONS • COMPLEX TRANSLOCATIONS • AND TELOMERE ALTERATIONS TRIPLE CHROMOSOME #20 A DELETION in CHROMOSOME #22 SPECTRAL KARYOTYPING CYTOGENETIC DISORDERS • DEFINITIONS: –EUPLOID (46XX or 46XY) –ANEUPLOID (NOT AN EXACT MULTIPLE OF 23) • MONOSOMY, AUTOSOME OR SEX • TRISOMY, AUTOSOME OR SEX –DELETION –BREAKAGE MORE DEFINITIONS COMMON CYTOGENETIC DISEASES • AUTOSOMES – TRISOMY-21 (DOWN SYNDROME) – 8, 9, 13 (Patau), 18 (Edwards), 22 – 22q.11.2 deletion • SEX CHROMOSOMES –KLINEFELTER: XXY, XXXY, etc. –TURNER: XO TRISOMY-21 TRISOMY-21 • Most trisomies (monosomies, aneuploidy) are from maternal non-disjunction • (non-disjunction or anaphase lag are BOTH possible) • #1 cause of mental retardation • Maternal age related • Congenital Heart Defects, risk for acute leukemias, GI atresias • Most LOVABLE of all God’s children Chromosome 22q11.2 Deletion Syndrome • Because of a DELETION, this cannot be detected by standard karyotyping and needs FISH • Cardiac defects, DiGeorge syndrome, velocardiofacial, CATCH* SEX CHROMOSOME DISORDERS • Problems related to sexual development and fertility • Discovered at time of puberty • Retardation related to the number of X chromosomes • If you have at least ONE “Y” chromosome, you are male KLINEFELTER (XXY, XXXY, etc.) • Hypogonadism found at puberty • #1 cause of male infertility • NO retardation unless more X’s • 47, XXY 82% of the time • L----O----N----G legs, atrophic testes, small penis TURNER (XO) • 45, X is the “proper” designation • Mosaics common • Often, the WHOLE chromosome is not missing, but just part • NECK “WEBBING” • EDEMA of HAND DORSUM • CONGENITAL HEART DEFECTS most FEARED • “STREAK” OVARIES HERMAPHRODITES ♂ ♀ • GENETIC SEX is determined by the PRESENCE or ABSENCE of a “Y” chromosome, but there is also, GONADAL (phenotypic), and DUCTAL sex • TRUE HERMAPHRODITE: OVARIES AND TESTES, often on opposite sides (VERY RARE) • PSEUDO-HERMAPHRODITE: – MALE: TESTES with female characteristics (Y-) – FEMALE: OVARIES with male characteristics (XX) SINGLE GENE, NON-Mendelian • Triplet repeats –Fragile X (CGG) –Others: ataxias, myotonic dystrophy • Mitochondrial Mutations: (maternal) (LEBER HEREDITARY OPTIC NEUROPATHY) • Genomic “IMPRINTING”: (Inactivation of maternal or paternal allele, contradicts Mendel) • Gonadal “MOSAICISM”: (only gametes have mutated cells) MOLECULAR DX by DNA PROBES • • • • • • • BIRTH DEFECTS, PRE- or POST- NATAL TUMOR CELLS CLASSIFICATIONS of TUMORS IDENTIFICATION of PATHOGENS DONOR COMPATIBILITY PATERNITY FORENSIC H&E tissue structures ImmunoAntigen Proteins GENES that MAKE those PROTEINS METHODS OF DNA ANALYSIS Fluorescence in Situ Hybridization (FISH) • FISH utilizes DNA probes that recognize sequences specific to chromosomal regions of greater than 100 kilobases in size, which defines the limit of resolution with this technique for identifying chromosomal changes. • Such probes are labeled with fluorescent dyes and applied to metaphase spreads or interphase nuclei. • The probe hybridizes to its complementary sequence on the chromosome and thus • labels the specific chromosomal region that can then be visualized under a fluorescence microscope. Array-Based Genomic Hybridization • FISH requires previous knowledge of the one or few specific chromosomal regions However, chromosomal abnormalities may also be detected without previous knowledge global strategy known as array based CGH. • Test DNA and a reference (normal) DNA are labeled with two different fluorescent dyes ( Cy5 and Cy3, which fluoresce red and green, ). The differentially labeled samples are then • hybridized to an array of segments of genomic • Amplifications and deletions in the test sample produce an increase or decrease in signal relative to the normal DNA that can be detected down to a 10-kilobase (kb) resolution • Newer generations of microarrays using • single-nucleotide polymorphisms (SNPs) provide even higher resolution Polymerase Chain Reaction (PCR) Analysis • Direct Detection of DNA Mutations by Polymerase Chain Reaction (PCR) Analysis PCR analysis, which involves exponential amplification of DNA, is now widely used in molecular diagnosis. • If RNA is used as the substrate, it is first reverse-transcribed to obtain cDNA and then amplified by PCR. This method involving reverse transcription (RT) often is abbreviated as RT-PCR. • One prerequisite for direct detection is that the sequence of the normal gene must be known. To detect the mutant gene, two primers that bind to the 3′ and 5′ ends of • the normal sequence are designed. By utilizing appropriate DNA polymerases and thermal cycling, the target DNA is greatly amplified, producing millions of copies of the DNA Linkage Analysis and GenomeWide Association Studies • Direct diagnosis of mutations is possible only if the gene responsible for a genetic disorder is known and its sequence has been identified. In several diseases that have a genetic • basis, including some common disorders, direct genetic diagnosis is not possible, either because the causal gene has not been identified or because the disease is multifactorial (polygenic) and no single gene is involved. • Two types of analyses can be performed for unbiased identification of disease-associated gene(s): • linkage analysis • genome-wide association studies (GWASs). In both surrogate markers in the genome, marker loci, must be used to localize the chromosomal regions of interest, based on their linkage to one regions • Prenatal genetic analysis should be offered to all patients who are at risk of having cytogenetically abnormal progeny. • It can be performed on cells obtained by amniocentesis, on chorionic villus biopsy material, or on umbilical cord blood . Indications are the following: Advanced maternal age (beyond 34 Years),which is associated with greater risk of trisomies Confirmed carrier status for a balanced reciprocal translocation, Robertsonian translocation, or inversion (inPrenatal genetic analysis should be offered to all patients • • A chromosomal abnormality affecting a previous child • • Determination of fetal sex when the patient or partner is a confirmed carrier of an X-linked genetic disorder • Postnatal genetic analysis usually is performed on peripheral blood lymphocytes. • • Multiple congenital anomalies • • Unexplained mental retardation and/or developmental delay • • Suspected aneuploidy (e.g., features of Down syndrome) • • Suspected unbalanced autosome (e.g., Prader-Willi syndrome)







