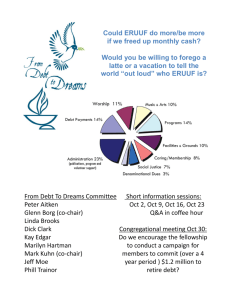
Presentation
advertisement

Visual Analytics and Biological Information Chris Shaw School of Interactive Arts & Technology, Simon Fraser University ______________________________________________________________________________________ SCHOOL OF INTERACTIVE ARTS + TECHNOLOGY [SIAT] | WWW.SIAT.SFU.CA Visual Analytics: Integrated Interdisciplinary R&D Information Systems Cognitive Science Visual Analytics Graphic & Interaction Design Oct 8, 2009 Mathematical & Statistical Methods 2 Interdisciplinary Know-how • SFU School of Interactive Arts & Tech – Design focus – Technology and Science – Cross-disciplinary Ph.D, MSc. BSc. • UBC Media & Graphics Interdisciplinary Centre (MAGIC) – 15 years use-inspired basic research,Co-development with industry & government 3 People Oct 8, 2009 IMAS: Interactive Multigenomic Analysis System 4 Visual Analytics • Two broad task domains: – Analysis of large datasets • Overall interaction time: hours to weeks • EG. the VAST Contest: Find the threat represented in this large collection of text, images, emails… – Monitoring and emergency response • Overall interaction time: seconds to minutes • EG. Airport security screening: Find the smuggled weapon Oct 8, 2009 5 NSERC Strategic Grant: “Visual Analytics for Safety & Security” • Application-aware basic research on humancomputer cognitive systems – Perceptual & spatial cognition stream Sensemaking, levels/types of user expertise New cognitive collaboration & coordination stream – Application development stream – – • Goal is to co-develop human & tech aspects – Understanding users, system customization and training is as important as new technology Oct 8, 2009 6 New Research Methods • Conventional qualitative and quantitative methods (e.g. grounded theory, stats) • Add advanced statistical, computational and math models • Integrated mixed-methods analysis • Calls for visual analytics tools for visual analytics research-- we are our testbed Oct 8, 2009 7 Goal is integrated Visual Analytics R&D improved understanding pure basic research existing understanding Oct 8, 2009 improved technology use-inspired basic research applied research and development existing technology 8 Science in the development process Walkthrough or experiment Design for key human information processing systems Interaction Science Assess specific aspects of interaction Implement prototype Oct 8, 2009 9 Steps From Research to Practice • VA science gives us design know-how • VA-aware designers work with user community to apply principles in design • VA users trained to get maximum advantage from VA • VA-sophisticated organizations work with designers to co-evolve new technology and work practices Oct 8, 2009 10 Biological Sequence Analysis • Visual Analytics in the domain of Biology – Large data – Non-spatial – Many different layers of abstraction Oct 8, 2009 11 Biological Sequence Analysis • DNA Sequencing projects • Visualization Systems • IMAS – Zoomable Sequence visualization – Gene Finding – BLAST Pairwise alignments – Multialignments • Results Oct 8, 2009 12 DNA Sequencing Projects • Similar sequence yields similar structure • Similar structure yields similar function Oct 8, 2009 13 IMAS supports • Initial stages of analyzing DNA sequence: – Find genes – Find and Analyze similar genes – Multialign like genes to find active sites • Pipeline structure Oct 8, 2009 14 Existing Tools • Typically web-based – Copy and paste sequence into text entry box – Await search or analysis on remote database – Get an isolated report that the user must organize • Visualization often done as a reporting function – UCSC Genome browser, LLNL ECR browser, NCBI annotation viewer Oct 8, 2009 15 Desktop Workbenches • Local sequence data – Mix of local and remote analyses • Web queries to remote data – Bluejay, Apollo, Vector NTI, CLC Workbench • User must work to integrate analyses • Workbench is point of collection Oct 8, 2009 16 IMAS • • • • Integrates analysis and display Horizontally Zoomable along sequence Selectable detail vertical Maintains a sequence analysis data collection • Visual display aligned to sequence Oct 8, 2009 17 IMAS Screenshot Oct 8, 2009 18 IMAS Contents • DNA Sequence (Nucleotide, or NT Seq) • GC % plot • 3 forward & 3 Reverse complement Amino Acid sequences: Oct 8, 2009 19 Genes • Built-in access to Glimmer 3.02 gene finder • The labelled boxes are anchors for sequence analysis • Segments of DNA can also be marked as a Feature for further analysis Oct 8, 2009 20 Analyses • Rricke104 gene has – 2 NT BLAST pairs – 1 AA BLAST pair – 1 NT multialignment – 1 AA multialignment Oct 8, 2009 21 Pairwise Sequence Analysis • Activated by selecting a Gene/Feature and selecting NT or AA similarity search • NCBI’s BLAST is called to search local databases of NT or AA sequences • Can also search NCBI central database Oct 8, 2009 22 BLAST Alignments • High Scoring Pairs are stacked from most to least significant score • Detail shown when zoomed in • Pair similarity is shown using background color – Darker blue indicates higher similarity • When zoomed out, text is hidden and only similarity is shown Oct 8, 2009 23 Multialignments • Select BLAST alignments to be multialigned • Clustal-W performs multialignment • Aligns – The originating IMAS gene sequence – The “Full” sequence found by BLAST • Not just the high-quality section – Useful to align entire genes, or entire corresponding segments of DNA Originating Gene BLAST Results Oct 8, 2009 24 Oct 30, 8, 2009 2007 IMAS: Interactive Multigenomic Analysis System 25 Results • Analyzed Orientia Tsutsugamushi (Scrub Typhus) – Found not much similarity in NT sequence – Found a large number of SMART domains not found in the related Rickettsia organisms • IMAS Benefit was data organization Oct 8, 2009 26 Discussion • Visualization Problems – Pair alignments need better organization • Local visibility and organization needed – Overlap in X causes stacking layout problems • Need selective relaxation of vertical alignment rule Oct 8, 2009 27 Discussion • Analysis Problems – More flexible access to tools: Restriction enzyme sites, methylation sites, Motifs, Primers, Transcription regulation, Intergenic signals...... • Database mediation problem: Please use XML! – More flexible manipulation of sequence parts • Right now IMAS is somewhat rigid in its worldview Oct 8, 2009 28 Multiple Genomes • Lots of organisms now sequenced: – Learn from individual similarities – Learn from similar gene organization – Co-location “Synteny” of genes helps infer similar function: • Located together -> expressed together Oct 8, 2009 29 Synteny Visualization • Line up the similar organisms below primary organism – Draw links to connect them – Take care to manage visual salience Oct 8, 2009 30 Synteny Visualization Final Oct 8, 2009 31 IMAS Synteny • Not so good with reversals: Oct 8, 2009 32 Alternative: Spring Synteny • Orthologs as a node-link diagram • 2 Link types – Neighbors on same organism – Sequence alignment (orthologous) links Oct 8, 2009 33 Alignment Links • Percent Identity Plot along sequence • Framed to show PIP range • RRickettsia linked to RConorii, RProwazekii, RTyphi, RAkari Oct 8, 2009 34 Springs • Primary organism is central spine – Secondary sequence have parallel track connected by similarity links – Each secondary sequence has its own resting length for similarity links • Length of neighbor links is blend of – NT coordinate difference – ln(length) * ln(length) Oct 8, 2009 35 Neighbor links • Using NT distance gives network shapes with many acute angles • Directly displays relative lengths of genomes Oct 8, 2009 36 Rrickettsii Genomes Oct 8, 2009 Genomic Spring-Synteny Visualization with IMAS 37 Results – Advantages: • Shows reversals clearly • Shows gene “splits” with respect to primary genome • Shows insertions/deletions – Disadvantages • Obscures length relationships • Force-directed layout requires fiddling • Rotating the similarity edge makes comparing similarity difficult Oct 30, 2007 38 Results • Trade-off: – Free 2 dimensions for gene placement • Get to locate similar items close to each other • Get ability to see gross rearrangements • Lose ability to see detailed similarity along DNA sequence • Lose geometric location information • Lose regulatory info (not represented) Oct 8, 2009 39 IMAS • Supports annotation pipeline • Tree or DAG visualization, where – Branches are individual BLAST runs – Branches converge on multialignments • Biologists want more! – Analyze arbitrary collections of sequence Oct 8, 2009 40 More • Want ability to interactively cut, edit, and analyze sequence • “Genomic Spreadsheet” where – Manage Sequences – Compare & Align sequences – Search for similar sequences – Manage sequences at levels of abstraction higher than sequence + annotation text Oct 8, 2009 41 CzSaw • A Visual Analytics System for Text Data • Built by the SIAT CzSaw group – Victor Chen, Dustin Dunsmuir, Nazanin Kadivar, Eric Lee, Cheryl Qian – John Dill, Chris Shaw, Rob Woodbury Oct 8, 2009 42 Exploring Data Analysis Visualizing Capturing Analysis Analysis Model Data Analysis Model Process Process Data Analysis History CzSaw Exploring Data Capturing Analysis Process Visualizing Analysis Model Analysis History Conclusions • Building IMAS helped us discover that IMAS is not yet what you want • Supports pipeline • Need to analyze with respect to many data types – Genome & other ontologies – Phylogeny – Metabolic networks – Regulatory networks Oct 8, 2009 50 Thanks! • Slides: Brian Fisher & John Dill • Greg Dasch, Marina Eremeeva, – Viral and Rickettsial Zoonoses Branch, CDC Atlanta Oct 8, 2009 51