1 2
advertisement

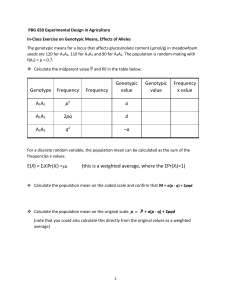
zygote viability selection survival to adult . courtship sexual selection fertilization sexual selection gamete production fecundity selection Fitness = individual’s genetic contribution to the next generation (zygotezygote); differential survival and/or reproduction absolute fitness, Wij = #offspring, lifespan, etc relative fitness, wij = contribution relative to other genotypes selection differential, sij = strength of selection against a genotype Pr(survival) Wij wij sij A1A1 A1A2 A2A2 80% 1.0 0 40% 0.5 0.5 20% 0.25 0.75 A numerical example Find the new genotype and allele frequencies A1A1 A1A2 A2A2 genotype freq. wij 0.25 1.0 0.50 0.75 0.25 0.25 Survival after selection 0.25(1) 0.5(0.75) 0.25 0.375 0.25(0.25) 0.0625 But what is the sum of these? 0.6875 To make them sum to one (for a new frequency) you must divide by 0.6875 What is 0.6875? It is the mean fitness. (p2w11 +2pqw12+q2w22) New genotype frequencies 0.363 What are the new allele frequencies? 0.546 0.091 A1 ~ 0.64 (0.5) A2 ~ 0.36 (0.5) w ’ = 0.363(1) + 0.546(0.75) + 0.091(0.25) = 0.7954 How does selection change genotype and allele frequencies? A1A1 A1A2 A2A2 p2 2pq q2 relative fitness, wij w11 w12 w22 geno. freq. after selection p2w11 w 2pqw12 w q2w22 w geno. freq. average relative fitness, w = p2w11 + 2pqw12 + q2w22 Patterns of selection -- Fitness arrays A1A1 w11 deleterious recessive recessive lethal deleterious dominant deleterious dominant deleterious intermediate deleterious recessive heterozygote advantage heteroz. disadvantage 1 1 1 1 1 1 1-s 1+s A1A2 w12 1 1 1 1-s 1-hs 1+s 1 1 A2A2 w22 1-s 0 1+s 1-s 1-s 1+s 1-t 1+t Selection against a recessive allele initial g.f. rel. fitness A1A1 A1A2 A2A2 P2 1 2pq 1 q2 1-s w = p2(1) + 2pq(1) + q2(1-s) = 1 – q2s g.f. > selection p2(1) 1-q2s 2pq(1) 1-q2s q2(1-s) 1-q2s A numerical example A1A1 A1A2 A2A2 gen. freq. 0.25 0.50 0.25 wij 1.0 1.0 0.4 gen. freq. > selection 0.25(1) 0.85 0.294 f’(A1) ~ 0.59 (0.5) 0.5(1) 0.85 0.588 0.25(0.4) 0.85 0.118 f’(A2) ~ 0.42 (0.5) w ’ = 0.294(1) + 0.588(1) + 0.25(0.4) = 0.982 what is the new frequency of A2 ? q’ = Q’ + 1 2 H’ pq q2(1-s) = + 2 1-sq2 1-sq q’ = q2(1-s) + pq 1-sq2 = q2 – sq2 + q – q2 1-sq2 = q(1-sq) 1-sq2 recall that p = 1 - q and q = 1 - p change in the frequency of a lethal recessive in Tribolium castaneum 2 change in the frequency of a deleterious recessive what is the new frequency of A2 ? q’ = Q’ + 1 2 H’ pq q2(1-s) = + 2 1-sq2 1-sq q’ = q2(1-s) + pq 1-sq2 = q2 – sq2 + q – q2 1-sq2 = q(1-sq) 1-sq2 recall that p = 1 - q and q = 1 - p how much has the frequency of A2 changed after one generation of selection ? Dq Dq = q’ - q = q(1-sq) 1-sq2 = q – sq2 – q + sq3 1-sq2 = -sq2(1 – q) 1-sq2 - q Dq selection against a deleterious recessive allele q Selection against a dominant allele initial g.f. rel. fitness A1A1 A1A2 A2A2 P2 1 2pq 1-s q2 1-s w = p2(1) + 2pq(1-s) + q2(1-s) = 1 – sq(2p-q) g.f. > selection p2(1) 1-sq(2p-q) 2pq(1-s) 1-sq(2p-q) q2(1-s) 1-sq(2p-q) change in the frequency of a deleterious dominant Selection against a dominant allele q Selection favoring heterozygotes initial g.f. rel. fitness A1A1 A1A2 A2A2 P2 1-s 2pq 1 q2 1-t w = p2(1-s) + 2pq(1) + q2(1-t) = 1 – p2s - q2t g.f. > selection p2(1-s) 2pq(1) 1 – p2s - q2t 1 – p2s - q2t q2(1-t) 1 – p2s - q2t q-tq2 Dq = 1-sp2-tq2 - q p-sp2 and Dp = - p 2 2 1-sp -tq at equilibrium, Dq = 0 and Dp = 0 q-tq2 w p-sp2 w =q 1 – tq w = 1 - sp w tq = sp = s(1-q) s q= s+t t p= s+t =p heterozygote advantage Change in allele frequency 0.10 0.05 0.00 -0.05 -0.10 0.0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 1.0 Allele frequency, q heterozygote advantage at phosphoglucose isomerase in Colias butterflies glycolysis enzyme kinetics of phosphoglucose isomerase in Colias deviation from expected heterozygosity .15 .10 .05 0 -.10 . 3 . . 11 17 July . -.05 23 Selection against heterozygotes initial g.f. rel. fitness A1A1 A1A2 A2A2 P2 1+s 2pq 1 q2 1+t w = p2(1+s) + 2pq(1) + q2(1+t) = 1 + p2s + q2t g.f. > selection p2(1+s) 1+p2s+q2t 2pq(1) 1+p2s+q2t q2(1+t) 1+p2s+q2t at equilibrium, Dq = 0 and Dp = 0 > s q = s+t and t p= s+t heterozygote disadvantage Change in allele frequency 0.10 0.05 0.00 -0.05 -0.10 0.0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 1.0 Allele frequency, q heterozygote disadvantage: translocation heterozygotes in Drosophila relative fitness of A1A1 simple models of selection w11 > w12 > w22 w11 > w12 < w22 fix A1 unstable polymorphism w11 < w12 > w22 w11 < w12 < w22 stable polymorphism fix A2 relative fitness of A2A2 relative fitness enables different traits and populations to be compared selection can act at many stages in the life cycle; opportunity for opposing selection at different stages directional selection fixes one allele and eliminates all others from the population heterozygote advantage can maintain a balanced polymorphism, but cannot explain the high levels of genetic variation found in natural populations heterozygote disadvantage produces an unstable polymorphism; which allele is fixed depends on chance