PROBLEM SET 8
advertisement

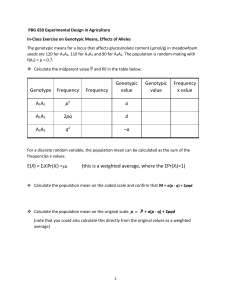
PROBLEM SET 7 POPULATION GENETICS The goal of this problem set is to add to your understanding of the regulation of phenotypic variation in populations. By reviewing some ideas in quantitative population genetics, you will explore the influence of various factors (dominance, allele frequency, and genotype values) on the frequency distribution of quantitative traits. The fundamental “theorem” of quantitative genetics is Vp = Vg + Ve, or phenotypic variance is the sum of genotypic and environmental variances. The exercise will consist of two parts. First, you will review theoretical expectations for distribution of phenotypes under various assumptions. Second, you will generate samples from populations using a Monte-Carlo technique. First, however some preliminaries. Assume the following genetic model for a quantitative trait: Y=G+E Genotype A1A1 A1A2 A2A2 Frequency p2 2pq q2 Mean Value a b c where Y is the phenotypic value, G is the genotype at an autosomal locus, and E is the random environmental effect; A1 and A2 are the allelles at the locus with gene frequencies p and q (p + q = 1). The quantities a, b, and c are mean values (e.g. size) of genotypes A1A1, A1A2, A2A2, having frequencies p2, 2pq and q2 respectively. Random environmental effects, E, are assumed to have a normal distribution with mean zero and an environmental variance, Ve. The probability density function of a phenotype Y would be: f(Y)= p[A1A1}*f(Y|A1A1) + p[A1A2]*f(Y|A1A2) + p[A2A2]*f(Y|A2A2) = p2*g(Y,a,Ve) + 2pq*g(Y,b,Ve) + q2*g(Y,c,Ve) 1 where g(Y,m,V) is a probability density function of a variable (Y) with mean m and variance V. The formula for the probability density function of a normal distribution, g(Y,a,Ve), is: Y a 2 1 gY , a ,Ve * exp . * Ve 2 * 31416 2 * Ve The parameters of this distribution are p, a, b, c and Ve. The following relationships may be useful for the evaluating the dependence of phenotypic distribution on its parameters: Overall genotypic mean: G = ap2 + 2bpq + cq2 Total genetic variance: Vg = a2p2 + 2b2pq + c2q2 - (G)2 = 2pq[p(a - b) + q(b - c)]2 + p2q2[a - 2b + c]2 = Va + Vd where Va is called the additive variance and Vd is called the variance due to dominance. Heritability: hg 2 Vg Vp and, Degree of dominance: x ha2 Va Vp a 2b c ac Part I: Set up a spread-sheet to calculate Vg, Va, Vd, Vp, ha, hg and x given a set of parameters values for p, a, b, c, and Ve. The spreadsheet should also contain a graph of f(Y) vs Y from at least 20 pairs of data. The range of Y should by plus or minus 2 standard deviations ( Ve ) beyond the maximum genotypic range. Evaluate the probability density function by varying parameters values to obtain three types of phenotypic distributions (trimodal, bimodal, and unimodal). Part II: Create a Visual Basic module to generate three sets of 500 phenotypes using these three sets of parameter values. Use our standard function to generate standard normal random variable to add environmental variability and random numbers to assign genotype of each of the 500 individuals. Remember our standard function is: R = 2* RND - 1 2 ZR = log ((1+R)/(1-R))/1.82 From each of three samples of 500 individuals, generate a histogram of frequency vs Y. The histograms should contain a minimum of 15 bins. SUBMIT: 1. An EXCEL file with a. Part I worksheet(s) and Part II module (3 points) b. Plots of 3 phenotypic distributions in Part I (5 points) c. Three histograms in Part II (5 points) 2. A document file with a. A flow chart of your Visual Basic module in Part II (3 points) b. A discussion comparing theoretical and simulated samples of frequencies of phenotypes. You should include insights you have gained about the influence of genotype values, dominance, and environmental variability on the observability of underlying genotypic frequencies in populations. You must elaborate on the values of Heritability and Dominance. Use your text book from Population Biology to discuss them. (24 points). 3