Conference program
advertisement

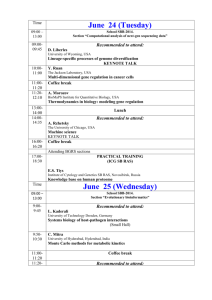
RUSSIAN ACADEMY OF SCIENCES SIBERIAN BRANCH INSTITUTE OF CYTOLOGY AND GENETICS PROGRAM OF THE FIFTH INTERNATIONAL CONFERENCE ON BIOINFORMATICS OF GENOME REGULATION AND STRUCTURE BGRS’2006 Novosibirsk, Russia July 16–22, 2006 Novosibirsk 2006 INTERNATIONAL PROGRAM COMMITTEE Nikolay Kolchanov Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia (Chairman of the Conference) Ralf Hofestaedt University of Bielefeld, Germany (Co-Chairman of the Conference) Dagmara Furman Institute of Cytology and Genetics SB RAS, Novosibirsk, (Conference Scientific Secretary) Dmitry Afonnikov Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia Mikhail Gelfand GosNIIGenetika, Moscow, Russia Vadim Govorun Institute of Physicochemical Medicine, RAMS, Moscow, Russia Reinhart Heinrich Humboldt University Berlin, Berlin, Germany Charlie Hodgman Multidisciplinary Centre for Integrative Biology, School of Biosciences, University of Nottingham, UK Alexey Kochetov Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia Eugene Koonin National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, USA Vassily Lyubetsky Institute for Informational Transmission Problems RAS, Moscow, Russia Luciano Milanesi National Research Council – Institute of Biomedical Technology, Italy Viatcheslav Mordvinov Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia Yuriy Orlov Genome Institute of Singapore, Singapore Igor Rogozin Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia Cenk Sahinalp Computing Science, Simon Fraser University, Burnaby, Canada Maria Samsonova St.Petersburg State Polytechnic University, St.Petersburg, Russia Akinori Sarai Kyushu Institute of Technology (KIT), Iizuka, Japan Konstantin Skryabin Centre "Bioengineering" RAS, Moscow, Russia Rustem Tchuraev Institute of Biology, Ufa Sci. Centre RAS, Ufa, Russia Denis Thieffry ESIL-GBMA, Universite de la Mediterranee, Marseille, France Jennifer Trelewicz IBM Almaden Research Center, San Jose, California, USA Edgar Wingender UKG, University of Goettingen, Goettingen, Germany Lev Zhivotovsky Institute of General Genetics RAS, Moscow, Russia Jagath C. Rajapakse School of Computer Engineering, Nanyang Technological University, Singapore LOCAL ORGANIZING COMMITTEE Sergey Lavryushev Institute of Cytology and Genetics, SB RAS, Novosibirsk (Chairperson) Ekaterina Denisova Institute of Cytology and Genetics, SB RAS, Novosibirsk Andrey Kharkevich Institute of Cytology and Genetics, SB RAS, Novosibirsk Galina Kiseleva Institute of Cytology and Genetics, SB RAS, Novosibirsk Anna Onchukova Institute of Cytology and Genetics, SB RAS, Novosibirsk Natalia Sournina Institute of Cytology and Genetics, SB RAS, Novosibirsk 2 Our sponsors Organizers Laboratory of Theoretical Genetics of the Institute of Cytology and Genetics, SB RAS Institute of Cytology and Genetics, SB RAS Siberian Branch of the Russian Academy of Sciences ВА В ИЛ О ВС КО Е О Б ЩЕС Т ВО ГЕНЕТИКОВ И СЕЛЕКЦИОНЕРОВ Vavilov Society of Geneticists and Breeders Scientific Council on Bioinformatics, Siberian Branch of the Russian Academy of Sciences Chair of Informational Biology, Department of Natural Sciences, Novosibirsk State University Grants INTAS Grant Russian Foundation for Basic Research 3 THE FIFTH INTERNATIONAL CONFERENCE ON BIOINFORMATICS OF GENOME REGULATION AND STRUCTURE (BGRS'2006) CONFERENCE PROGRAM SCHEDULE AT A GLANCE The Conference Sessions will be held in the Small Conference Hall and in Music Salon (room no. 220) of the House of Scientists July 16, Sunday Foyer of the Small Conference Hall 10:00-14:00 Registration of the conference participants* *Participants who come later are welcome for registration in the Organizing Committee room (House of Scientists, room no.200) on any conference day. 12:30-14:00 Lunch 14:00 Small Conference Hall Conference Opens: Welcome by Academician of the Russian Academy of Sciences Vladimir Shumny, Director of the Institute of Cytology and Genetics, Siberian Branch of the Russian Academy of Sciences Greetings of Corresponding member of the Russian Academy of Sciences, Professor Nikolay Kolchanov and Professor Ralf Hofestaedt 14:30-18:00 PLENAR SESSION 16:00-16:30 Coffee break 19:00-23:00 Welcome party July 17, Monday Small Conference Hall 9:30-12:45 Morning Session COMPUTATIONAL STRUCTURAL AND FUNCTIONAL GENOMICS 10:40-11:10 Coffee break 12:45-14:30 Lunch break 14:30-17:45 Evening Session COMPUTATIONAL STRUCTURAL AND FUNCTIONAL GENOMICS 16:00-16:30 Coffee break Computer demos of this session will be available during the entire conference day July 18, Tuesday Small Conference Hall 9:30-12:50 Morning Session COMPUTATIONAL STRUCTURAL AND FUNCTIONAL PROTEOMICS 10:45-11:15 Coffee break 12:50 -14:30 Lunch break 4 14:30 -17:25 Evening Session COMPUTATIONAL STRUCTURAL AND FUNCTIONAL PROTEOMICS 16:05-16:35 Coffee break Computer demos of this session will be available during the entire conference day July 19, Wednesday Foyer of the Small Conference Hall 9:30-12:30 POSTER SESSION 10:45-11:15 Coffee break 12:30-14:00 Lunch break July 20, Thursday Small Conference Hall 9:30-12:55 Morning Session COMPARATIVE AND EVOLUTIONARY GENOMICS AND PROTEOMICS 10:45-11:15 Coffee break 12:55-14:30 Lunch break 14:30-18:10 Evening Session COMPARATIVE AND EVOLUTIONARY GENOMICS AND PROTEOMICS 16:00-16:30 Coffee break Computer demos of this session will be available during the entire conference day July 20, Thursday Music Salon (room no.220) 9:30-12:55 Morning Session NEW APPROACHES TO BIOMOLECULAR DATA AND PROCESSES ANALYSIS/MODELING 10:45-11:15 Coffee break 12:55-14:30 Lunch break 14:30-17:20 Evening Session NEW APPROACHES TO BIOMOLECULAR DATA AND PROCESSES ANALYSIS/MODELING 16:05-16:30 Coffee break July 21, Friday Small Conference Hall 9:30-12:55 Morning Session COMPUTATIONAL SYSTEMS BIOLOGY 10:45-11:15 Coffee break 12:50-14:30 Lunch break Computer demos of this session will be available during the entire conference day 5 July 22, Saturday Small Conference Hall 9:30-12:50 Morning Session COMPUTATIONAL SYSTEMS BIOLOGY 10:45-11:15 Coffee break 12:50-14:30 Lunch break 14:30-16:40 Evening Session COMPUTATIONAL SYSTEMS BIOLOGY 15:20-15:50 Coffee break Computer demos of this session will be available during the entire conference day 16:40-17:20 Closing Ceremony; Comments of Session Chairpersons PLENAR SESSION July 16, Sunday House of Scientists, Small Conference Hall 14:30-18:00 Co-chairpersons: Prof. Ralf Hofestaedt, Bielefeld University, AG Bioinformatics, Bielefeld, Germany Prof. Nikolay Kolchanov, Institute of Cytology and Genetics, Siberian Branch of the Russian Academy of Sciences, Novosibirsk, Russia No. 1. 2. Author(s) and Title of Talk Mazo I. *, Sivachenko A., Yuryev A., Daraselia N. Ariadne Genomics, Inc., Rockville, USA MOLECULAR NETWORKS IN MAMMALS: EXTRACTION FROM LITERATURE AND MICROARRAY Rodin S.N.1*, Rodin A.S.2 1 Beckman Research Institute of the City of Hope, Duarte, CA, USA 2 Human Genetics Center, School of Public Health, University of Texas, Houston, TX, USA GENE DUPLICATIONS IN EVOLUTION: FROM ORIGIN OF THE GENETIC CODE TO THE HUMAN GENOME Timeline 14:3015:15 15:1516:00 Coffee/tea break 16:00-16:30 3. 4. Ivanov A.S.*, Gnedenko O.V., Molnar A.A., Mezentsev Yu.V., Lisitsa A.V., Archakov A.I. V.N.Orechovich Institute of Biomedical Chemistry RAMS, Moscow, Russia PROTEIN-PROTEIN INTERACTIONS AS NEW TARGETS FOR DRUG DESIGN: INTERACTIVE LINKS BETWEEN VIRTUAL AND EXPERIMENTAL APPROACHES Samsonova M.G.1, Reinitz J.2 1 St. Petersburg State Polytechnical University, St. Petersburg, Russia 2 Stony Brook University, Stony Brook, New York, USA SYSTEMS BIOLOGY OF SEGMENT DETERMINATION 6 16:3017:15 17:1518:00 ORAL PRESENTATIONS Dear speakers, please, plan your presentation time including 5 minutes for questions. Only PowerPoint presentations made in Microsoft Office 2003 or earlier versions can be accepted. Use embedded pictures and animations only; picture and animation links to the Internet or to other files will not be accessible. Slide projectors or overhead projectors are NOT available during the BGRS'2006 Conference. Please, take into account that it will not be possible for you to load your presentation during the session by yourselves. Presentations are loaded on the main computer by the technician of the Organizing Committee only. To ensure that the presentations are well prepared and compatible with the main presentation computer, we strongly advice you to bring your materials as PowerPoint files on USB device or CD disk to the Room no. 200 at the House of Scientists and check the presentation with the technician of the Organizing Committee not later than 30 min before the start time of the session where you are presenting. This will prevent any confusion when loading the presentations on the main computer. You may also bring your laptop for copying your presentation to the main presentation computer. Use of your own laptop during your presentation is not recommended. If you have any questions, please do not hesitate to contact the Organizing Committee in Room no. 200 at the House of Scientists. 7 July 17, Monday House of Scientists, Small Conference Hall Morning Session 9:30-12:45 COMPUTATIONAL STRUCTURAL AND FUNCTIONAL GENOMICS AND TRANSCRIPTOMICS Co-chairpersons: Prof. Andrey Mironov, Department of Bioengineering and Bioinformatics, Moscow State University, Moscow, Russia Dr. Georgii Bazykin, Department of Ecology and Evolutionary Biology, Princeton University, Princeton, NJ, USA No. 1. 2. 3. Author(s) and Title of Talk Seliverstov A.V.*, Lyubetsky V.A. Institute for Information Transmission Problems RAS, Moscow, Russia GENE EXPRESSION REGULATION IN ACTINOBACTERIA AND CHLOROPLASTS Grosse I. 1,2 1 Leibniz Institute of Plant Genetics and Crop Plant Research (IPK) 2 Bioinformatics Center Gatersleben-Halle, Gatersleben, Germany COMPUTATIONAL IDENTIFICATION OF TRANSCRIPTION FACTOR BINDING SITES WITH VARIABLE ORDER BAYESIAN NETWORKS Levitsky V.G.1,2*, Ignatieva E.V.1,2, Ananko E.A.1, Merkulova T.I.1 1 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia 2 Novosibirsk State University, Novosibirsk, Russia THE SITEGA AND PWM METHODS APPLICATION FOR TRANSCRIPTION FACTOR BINDING SITES RECOGNITION IN EPD PROMOTERS Timeline 9:309:55 9:5510:20 10:2010:40 Coffee/tea break 10:40-11:10 4. 5. 6. 7. Shelest E.S.*1, Wingender E.1,2 1 Department of Bioinformatics, UKG, University of Göttingen, Göttingen, Germany 2 BIOBASE GmbH, Wolfenbüttel, Germany PROMOTER MODELING APPROACHES APPLIED TO THE INVESTIGATION OF p63 UP- AND DOWNSTREAM PROMOTERS Vorobjev Y.N., Emelianov D.Y. Institute of Chemical Biology and Fundamental Medicine, SB RAS, Novosibirsk, Russia MODELING OF DATA BASE OF CONTEXT-DEPENDENT CONFORMATIONAL PARAMETERS OF DNA DUPLEXES Kamzolova S.G., Osypov A.A.*, Dzhelyadin T.R., Beskaravainy P.M., Sorokin A.A. Institute of Cell Biophysics of RAS, Pushchino, Moscow region, Russia CONTEXT-DEPENDENT EFFECTS OF UPSTREAM A-TRACTS ON PROMOTER ELECTROSTATIC PROPERTIES AND FUNCTION Vishnevsky O.V.1,2*, Konstantinov Yu.M.3 1 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia 2 Novosibirsk State University, Novosibirsk, Russia 3 Siberian Institute of Plant Physiology and Biochemistry SB RAS, Irkutsk, Russia ANALYSIS OF THE NUCLEOTIDE CONTEXT OF HIGHER PLANT MITOCHONDRIAL mRNA EDITING SITES 12:45-14:30 Lunch break 8 11:1011:35 11:3512:00 12:0012:20 12:2012:45 July 17, Monday House of Scientists, Small Conference Hall Evening Session 14:30-18:10 COMPUTATIONAL STRUCTURAL AND FUNCTIONAL GENOMICS AND TRANSCRIPTOMICS Co-chairpersons: Prof. Vasily Lyubetsky, Institute for Information Transmission Problems, the Russian Academy of Sciences, Moscow, Russia Dr. Ivo Grosse, Leibniz Institute of Plant Genetics and Crop Plant Research (IPK) and Bioinformatics Center Gatersleben-Halle, Gatersleben, Germany No. 1. 2. 3. 4. Author(s) and Title of Talk 1 1 Timeline 1,2 1,2 Omelianchuk N.A. , Khomicheva I.V. , Savinskaya S.A. , Levitsky V.G. , Vishnevsky O.V. 1,2, Katokhin A.V.1,2, Ponomarenko M.P.1 , Kolchanov N.A.1,2 1 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia 2 Novosibirsk State University, Novosibirsk Russia REVEALING THE FUNCTIONAL-SPECIFIC TEXTUAL FEATURES OF THE microRNA SEQUENCES IN ARABIDOPSIS THALIANA Tutukina M.N., Masulis I.S., Ozoline O.N.* Institute of Cell Biophysics RAS, Pushchino, Moscow region, Russia TOWARDS THE IDENTIFICATION OF ANTISENSE RNA WITHIN GENES OF TRANSCRIPTION REGULATORS Orlov Yu.L.1, Zhou J.1, Lipovich L.1, Yong H.C.2, Li Yi2, Shahab A.2, Kuznetsov V.A.1* 1 Genome Institute of Singapore, Singapore 2 Bioinformatics Institute, Singapore A COMPREHENSIVE QUALITY ASSESSMENT OF THE AFFYMETRIX U133A/B PROBESETS BY AN INTEGRATED GENOMIC AND CLINICAL APPROACH Kuznetsov V.A.*, Zhou J., George J., Orlov Yu.L. Genome Institute of Singapore, Singapore GENOME-WIDE CO-EXPRESSION PATTERNS OF HUMAN CIS-ANTISENSE GENE PAIRS 14:3014:55 14:5515:15 15:1515:35 15:3516:00 Coffee/tea break 16:00-16:30 5. 6. 7. Efimov V.M.1*, Badratinov M.S.2, Katokhin A.V.2,3 1 Institute of Systematics and Ecology of Animals, SB RAS, Novosibirsk, Russia 2 Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia 3 Novosibirsk State University, Novosibirsk, Russia INTERPRETATION OF RESULTS OF SOM ANALYSIS OF MICROARRAY DATA BY PRINCIPAL COMPONENTS Subkhankulova T., Livesey F.J. Gurdon Institute and Department of Biochemistry, University of Cambridge, Tennis Court Road, Cambridge, CB2 1 QN, UK HOW SIMILAR ARE PHENOTYPICALLY IDENTICAL CELLS AT THE TRANSCRIPTIONAL LEVEL Abhishek K.1*, Karnick H.1, Mitra P.2 1 Indian Institute of Technology Kanpur, Kanpur, India 2 Indian Institute of Technology Kharagpur, Kharagpur, India A KNOWLEDGE AND DATA BASED HYBRID APPROACH TO GENE CLUSTERING 9 16:3016:55 16:5517:20 17:2017:45 July 18, Tuesday House of Scientists, Small Conference Hall Morning Session 9:30-12:50 COMPUTATIONAL STRUCTURAL AND FUNCTIONAL PROTEOMICS Co-chairpersons: Dr. Kaiser Jamil, Indo American Cancer Institute and Research Center, Hyderabad, India Dr. Roman Efremov, M.M. Shemyakin and U.A.Ovchinnikov Institute of Bioorganic Chemistry, the Russian Academy of Sciences, Moscow, Russia No. 1. 2. 3. Author(s) and Title of Talk Vlasov P.K.*, Esipova N.G., Tumanyan V.G. Engelhardt Institute of Molecular Biology, Russian Academy of Sciences, Moscow, Russia CONFORMATION PROPERTIES OF SHORT OLIGOPEPTIDES AND PREDICTION OF PROTEIN CHAIN CONFORMATION Volynsky P.E.*, Vereshaga Ya. A., Nolde D.E., Efremov R.G. M.M.Shemyakin and Yu.A.Ovchinnikov Institute of Bioorganic Chemistry RAS, Moscow, Russia PROBING DIMERIZATION OF TRANSMEMBRANE PEPTIDES VIA MOLECULAR DYNAMICS IN EXPLICIT BILAYERS Chekmarev S.F.1*, Krivov S.V.2, Karplus M.2,3* 1 Institute of Thermophysics SB RAS, Novosibirsk, Russia 2 Laboratoire de Chimie Biophysique, ISIS, Université Louis Pasteur, Strasbourg, France 3 Department of Chemistry and Chemical Biology, Harvard University, Cambridge, MA, USA “STRANGE KINETICS” OF UBIQUITIN FOLDING: INTERPRETATION IN TERMS OF A SIMPLE KINETIC MODEL Timeline 9:309:55 9:5510:20 10:2010:45 Coffee/tea break 10:45-11:15 4. 5. 6. 7. Afonnikov D.A.1,2 1 Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia 2 Novosibirsk State University, Novosibirsk, Russia RESIDUE-RESIDUE CONTACT MAP PREDICTION FOR PROTEINS Aksianov E.1*, Zanegina O.2, Alexeevski A.3, Karyagina A.4,5, Spirin S.3 1 Virology Department, Biological Faculty, Moscow State University, Moscow, Russia 2 Bioengineering and Bioinformatics Faculty, Moscow State University, Moscow, Russia 3 Belozersky Institute, Moscow State University, Moscow, Russia 4 N.F.Gamaleya Research Institute of Epidemiology and Microbiology, Moscow, Russia 5 Institute of Agricultural Biotechnology, Moscow, Russia A TOOL FOR COMPARATIVE ANALYSIS OF SOLVENT MOLECULES IN PDB STRUCTURES Naumoff D.G. State Institute for Genetics and Selection of Industrial Microorganisms, Moscow, Russia DEVELOPMENT OF A HIERARCHICAL CLASSIFICATION OF THE TIM-BARREL TYPE GLYCOSIDE HYDROLASES Safro M.*1,Tworowski D.1, Feldman A.2 1 Department of Structural Biology, Weizmann Institute of Science, Rehovot, Israel 2 EML Research gGmbH, Heidelberg, Germany HOW ARE CHARGED RESIDUES DISTRIBUTED AMONG FUNCTIONALLY DISTINCT STRUCTURAL DOMAINS OF AMINOACYL-tRNA SYNTHETASES? 12:50-14:30 Lunch break 10 11:1511:40 11:4012:05 12:0512:25 12:2512:50 July 18, Tuesday House of Scientists, Small Conference Hall Evening Session 14:30-17:25 COMPUTATIONAL STRUCTURAL AND FUNCTIONAL PROTEOMICS Co-chairpersons: Dr. Vladimir Kuznetsov, Genome Institute of Singapore, Singapore Prof. Sergei Chekmarev, Institute of Thermophysics Siberian Branch of the Russian Academy of Sciences, Novosibirsk, Russia No. 1. 2. 3. 4. Author(s) and Title of Talk I.S. 3, Korotkov R.O. 3, 1 1,3 3 Aman E.E. , Pintus S.S. , Startcev K.S. , Tatarnikova L.Yu. 3, Panina I.I. 3, Sobolev A.A. 3, Seleznev I.V. 3, Elohov V.Yu. 3, Golovina A.N. 3, Aknazarov Z.I. 3, Sharonova I.V. 3, Krestianova M.A. 3, Mishchenko E.L. 1, Magdysuk A.V. 3, Nemiatov A.I. 3 1 Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia 2 Institute of Mathematics SB RAS, Novosibirsk, Russia 3 Novosibirsk State University, Novosibirsk, Russia FASTPROT: A COMPUTATIONAL WORKBENCH FOR ANALYSIS OF FUNCTION, ACTIVITY AND STRUCTURE OF PROTEINS Chernorudskiy A.L., Shorina A.S., Garcia A., Gainullin M.R.* Nizhny Novgorod State Medical Academy, Nizhny Novgorod, Russia DIRECT INFLUENCE OF UBIQUITYLATION ON A TARGET PROTEIN ACTIVITY: “LOSS-OF-FUNCTION” MECHANISM REVEALED BY COMPUTATIONAL ANALYSIS Demenkov P.S.1*, Aman E.E.2, Ivanisenko V.A.2,3 1 Sobolev Institute of Mathematics of the SB RAS, Novosibirsk, Russia 2 Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia 3 Novosibirsk State University, Novosibirsk, Russia PREDICTION IN CHANGES OF PROTEIN THERMODYNAMIC STABILITY UPON SINGLE MUTATIONS Kochetov A.V.1*, Sarai A.2, Kolchanov N.A.1 1 Institute of Cytology and Genetics, Novosibirsk, Russia 2 Kyushu Institute of Technology, Dept. Biochemical Engineering and Science, Iizuka, Japan THE CONTRIBUTION OF ALTERNATIVE TRANSLATION START SITES TO HUMAN PROTEIN DIVERSITY Timeline Ivanisenko V.A. 1, Demenkov P.S. 2, Fomin E.S. 1, Oshurkov 14:3014:55 14:5515:20 15:2015:40 15:4016:05 Coffee/tea break 16:05-16:35 5. 6. Efremov R.G. Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry RAS, Moscow, Russia COMPUTER SIMULATIONS OF PEPTIDES AND PROTEINS IN MEMBRANES: PERSPECTIVES IN DRUG DESIGN Nicklaus M. Center for Cancer Research, NCI/NIH, Frederick, USA NEW DEVELOPMENTS AT THE NIH FOR INTEGRATION OF SMALLMOLECULE DATABASES AND BIOINFORMATICS RESOURCES 11 16:3517:00 17:0017:25 July 20, Thursday House of Scientists, Small Conference Hall Morning Session 9:30-12:55 COMPARATIVE AND EVOLUTIONARY GENOMICS AND PROTEOMICS Co-chairpersons: Dr. Vladimir Kapitonov, Genetic Information Research Institute, Mountain View, California, USA Dr. Igor Makunin, ARC Special Research Centre for Functional and Applied Genomics, Institute for Molecular Bioscience, University of Queensland, Brisbane, Australia No. 1. 2. 3. Author(s) and Title of Talk Vitreschak A.G.1*, Lyubetsky V.A.1, Gelfand M.S.1 1 Institute for Information Transmission Problems of RAS, Moscow, Russia EVOLUTIONAL AND FUNCTIONAL ANALYSIS OF T-BOX REGULON IN BACTERIA. IDENTIFICATION AND CHARACTERIZATION OF NEW GENES INVOLVED IN AMINO ACID METABOLISM Bazykin G.A.1*, Dushoff J.1, Levin S.1, Kondrashov A.2 1 Department. of Ecology and Evolutionary Biology, Princeton University, Princeton, NJ, USA 2 National Center for Biotechnology Information, NIH, Bethesda, USA INSTANCES OF POSITIVE SELECTION AT CONSERVATIVE PROTEIN Gunbin K.V., Suslov V.V., Kolchanov N.A.* Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia AROMORPHOSES AND ADAPTIVE MOLECULAR EVOLUTION: MORPHOGENS AND SIGNALING CASCADE GENES Timeline 9:309:55 9:5510:20 10:2010:45 Coffee/tea break 10:45-11:15 4. 5. 6. 7. Kondrashov F.A.1*, Koonin E.V.2, Morgunov I.G.3, Finogenova T.V.3, Kondrashova M.N.4 1 Section on Ecology, Behavior and Evolution, Division of Biological Sciences, University of California at San Diego, La Jolla, USA 2 National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, USA 3 Skryabin Institute of Biochemistry and Physiology of Microorganisms RAS, Pushchino, Russia 4 Institute of Theoretical and Experimental Biophysics RAS, Pushchino, Russia EVOLUTION OF THE GLYOXYLATE CYCLE ENZYMES IN THE METAZOA Ramensky V.1*, Nurtdinov R.2, Neverov A.3, Mironov A. 2, Gelfand M. 2,4 1 Engelhardt Institute of Molecular Biology RAS, Moscow, Russia 2 Department of Bioengineering and Bioinformatics, Moscow State University, Moscow, Russia 3 State Scientific Center GosNIIGenetika, Russia 4 Institute for Information Transmission Problems, RAS, Moscow, Russia HUMAN GENOME POLYMORPHISM AND ALTERNATIVE SPLICING Pintus S.S.1*, Fomin E.S.1, Oshurkov I.S.2, IvanisenkoV.A.1,2* 1 Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia 2 Novosibirsk State University, Novosibirsk, Russia PHYLOGENETIC ANALYSIS OF THE p53 AND P63/P73 GENE FAMILIES Sabitha K. 1, 2, Jamil K.1, 2 * 1 Genetics Department, Bhagawan Mahavir Hospital and Research Center, Hyderabad, India 2 Indo American Cancer Institute and Research Center, Hyderabad, India ANALYSIS OF EGFR GENE MUTATIONS WHICH HAVE A RESPONSE TO QUINAZOLIN INHIBITORS 12:55-14:30 Lunch break 12 11:1511:40 11:4012:05 12:0512:30 12:3012:55 July 20, Thursday House of Scientists, Small Conference Hall Evening Session 14:30-18:10 COMPARATIVE AND EVOLUTIONARY GENOMICS AND PROTEOMICS Co-chairpersons: Dr. Sergei Rodin, Beckman Research Institute of the City of Hope, Duarte, USA Dr. Vassily Ramensky, Engelhardt Institute of Molecular Biology, the Russian Academy of Sciences, Moscow, Russia No. 1. 2. 3. 4. Author(s) and Title of Talk Ignatieva E.V.*, Oshchepkov D.Yu., Klimova N.V., Vasiliev G.V., Merkulova T.I. Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia ANALYSIS OF REGULATORY REGIONS OF ORTHOLOGOUS GENES: PREDICTION OF SF1 BINDING SITES WITH SITECON METHOD PROVED BY EXPERIMENTAL VERIFICATION AND COMPARATIVE GENOMICS Kolesnikov N.N.*, Elisafenko E.A., Zakian S.M. Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia EVOLUTION OF THE STRUCTURE OF THE XIST LOCUS IN MAMMALS Deyneko I.V.*1,2, Kalybaeva Y.M.1, Kel A.E. 3, Blöcker H.*1, Kauer G. 4 1 Department of Genome Analysis, GBF, Braunschweig, Germany 2 Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia 3 BIOBASE GmbH, Wolfenbüttel, Germany 4 University of Applied Sciences, Emden, Germany HUMAN-CHIMPANZEE PROPERTY-DEPENDANT COMPARISONS ON CHROMOSOMES 21 Perelygin A.A.*1, Zharkikh A.A.2, Brinton M.A.1 1 Biology Department, Georgia State University, Atlanta, GA, USA 2 Bioinformatics Department, Myriad Genetics, Inc., Salt Lake City, UT, USA CONCERTED EVOLUTION OF PARALOGOUS Oas1 GENES IN RODENTIA AND CETARTIODACTYLA Timeline 14:3014:55 14:5515:20 15:2015:35 15:3516:00 Coffee/tea break 16:00-16:30 5. 6. 7. 8. Kapitonov V.V.*, Jurka J. Genetic Information Research Institute, Mountain View, California, USA NEW KIDS ON THE BLOCK: SELF-SYNTHESIZING DNA TRANSPOSONS Simons C., Pheasant M., Makunin I.V., Mattick J.S.* ARC Special Research Centre for Functional and Applied Genomics, Institute for Molecular Bioscience, University of Queensland, Brisbane, Australia TRANSPOSON-FREE REGIONS IN MAMMALIAN GENOMES Golovnina K.1, Glushkov S.*1, Blinov A.1, Mayorov V.2, Adkison L.2 , Goncharov N.1 1 Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia 2 Mercer University School of Medicine, Macon, USA MOLECULAR PHYLOGENY OF THE GENUS TRITICUM L. Karafet T. M.1,2*, Lansing J.S.2, Hammer M.F.2 1 Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia 2 University of Arizona, Tucson, USA IN SEARCH OF GENETIC SIGNATURE FOR THE EXPANSION OF IRRIGATION SYSTEMS IN BALI 13 16:3016:55 16:5517:20 17:2017:45 17:4518:10 July 20, Thursday House of Scientists, Music Salon (room no. 200) Morning Session 9:30-12:55 NEW APPROACHES TO BIOMOLECULAR DATA AND PROCESSES ANALYSIS/MODELING Co-chairpersons: Prof. Mikhail Lavrentiev, Jr., Novosibirsk State University, Novosibirsk, Russia Prof. Vladimir Golubyatnikov, Institute of Mathematics, Siberian Branch of the Russian Academy of Sciences, Novosibirsk, Russia No. 1. 2. 3. Author(s) and Title of Talk Mironov A.A.1,2,3 1 Department of Bioengineering and Bioinformatics, Moscow State University, Moscow, Russia 2 Institite for Information Transmission Problems, RAS, Moscow, Russia 3 State Scientific Center GosNIIGenetica, Russia THRESHOLD SELECTION USING THE RANK STATISTICS Hofestaedt R. Bielefeld University, AG Bioinformatics, Bielefeld, Germany AN EXTENDED BACKUS-SYSTEM FOR THE REPRESENTATION AND ANALYSIS OF DNA SEQUENCES Mjolsness E. Institute for Genomics and Bioinformatics and Departments of Computer Science and Mathematics, University of California, Irvine, USA MODELING TRANSCRIPTIONAL REGULATION WITH EQUILIBRIUM MOLECULAR COMPLEX COMPOSITION Timeline 9:309:55 9:5510:20 10:2010:45 Coffee/tea break 10:45-11:15 4. 5. 6. 7. Matveeva A.*, Kozlov K., Samsonova M.G. Department of Computational Biology, Center for Advanced Studies St. Petersburg State Polytechnical University, St. Petersburg, Russia METHODOLOGY FOR BUILDING OF COMPLEX WORKFLOWS WITH PROSTAK PACKAGE AND ISIMBIOS Tatarinova T.1, Schumitzky A.2 1 Ceres, Inc. Thousand Oaks, California, USA 2 University of Southern California, Los Angeles, USA MULTIPLE COLLAPSE CLUSTERING Litvinov I. I.1,3*, Finkelshtein A. V.2, Roytberg M. A.1,3* 1 Institute of Mathematical Problems in Biology, Russian Academy of Sciences, Pushchino, Moscow region, Russia 2 Institute of Protein Research, Russian Academy of Sciences, Pushchino, Moscow region, Russia 3 Pushchino State University, Pushchino, Moscow region, Russia OPTIMIZATION OF ACCURACY AND CONFIDENCE FOR ALIGNMENT ALGORITHMS EXPLOITING DATA ON SECONDARY STRUCTURE Kozlov K.N.*1, Samsonov A.M.2 1 Department of Computational Biology, State Polytechnical University, St.Petersburg, Russia 2 A.F. Ioffe Physico-technical Institute of the Russian Academy of Sciences, St.Petersburg, Russia NEW MIGRATION SCHEME FOR PARALLEL DIFFERENTIAL EVOLUTION 12:55-14:30 Lunch break 14 11:1511:40 11:4012:05 12:0512:30 12:3012:55 July 20, Thursday House of Scientists, Music Salon (room no. 200) Evening Session 14:30-17:20 NEW APPROACHES TO BIOMOLECULAR DATA AND PROCESSES ANALYSIS/MODELING Co-chairpersons: Prof. Mikhail Roytberg, Institute of Mathematical Problems in Biology, the Russian Academy of Sciences, Pushchino, Moscow Region, Russia Dr. Andrei Zinovyev, Institute Curie, Bioinformatics Service, Paris, France No. 1. 2. 3. 4. Author(s) and Title of Talk Demidenko G.V.*, Matveeva I.I. Sobolev Institute of Mathematics, SB RAS, Novosibirsk, Russia ASYMPTOTIC PROPERTIES OF SOLUTIONS OF DIFFERENTIALDIFFERENCE EQUATIONS WITH PERIODIC COEFFICIENTS IN LINEAR TERMS Mateus D.*, Gallois J.P. CEA/LIST Saclay, F-91191 Gif sur Yvette Cedex, France SEARCHING CONSTRAINTS IN BIOLOGICAL REGULATORY NETWORKS USING SYMBOLIC ANALYSIS Labarga A.*, Anderson M., Valentin F., Lopez R. European Bioinformatics Institute, Hinxton, United Kingdom WEB SERVICES AT THE EUROPEAN BIOINFORMATICS INSTITUTE Kolpakov F.1,2*, Puzanov M.1,2, Koshukov A.1,2 1 Institute of Systems Biology OOO, Novosibirsk, Russia 2 Design Technological Institute of Digital Techniques SB RAS, Novosibirsk, Russia BioUML: VISUAL MODELING, AUTOMATED CODE GENERATION AND SIMULATION OF BIOLOGICAL SYSTEMS Timeline 14:3014:55 14:5515:20 15:2015:45 15:4516:05 Coffee/tea break 16:05-16:30 5. 6. Miginsky D.S.1,2*, Suslov V.V.1, Rasskazov D.A.1,2, Podkolodny N.L.1, Kolchanov N.A.1,2 1 Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia 2 Novosibirsk State University, Novosibirsk, Russia ARCHITECTURE OF SOFTWARE TOOLKIT FOR STORING AND OPERATNG WITH BIOSYSTEMS MODELS Lohmann M. Cologne University Bioinformatics Center (CUBIC), Cologne, Germany METABOLOMICS, BIOINFORMATICS, SYSTEMS BIOLOGY — AN OVERVIEW OF RESEARCH AND TEACHING ACTIVITIES AT CUBIC 15 16:3016:55 16:5517:20 July 21, Friday House of Scientists, Small Conference Hall Morning Session 9:30-12:55 COMPUTATIONAL SYSTEMS BIOLOGY Co-chairpersons: Prof. Eric Mjolsness, Institute for Genomics and Bioinformatics and Departments of Computer Science and Mathematics, University of California, Irvine, USA Dr. Bjorn Olsson, School of Humanities and Informatics, University of Skövde, Sweden No. 1. 2. 3. Author(s) and Title of Talk Golubyatnikov Gaidov Yu.A.2, Kleshchev A.G.1, Volokitin E.P.1 1 Institute of Mathematics, SB RAS, Novosibirsk, Russia 2 Novosibirsk State University, Novosibirsk, Russia A GENE NETWORK MODEL WITH DIFFERENT TYPES OF REGULATION Likhoshvai V.A. 1 Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia MOLECULAR-GENETIC SYSTEMS: SYMMETRY OF STRUCTURE AND DYNAMIC CHAOS Khlebodarova T.M.*, Kachko A.V., Stepanenko I.L., Tikunova N.V. Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia DATABASE GenSensor AS INFORMATIONAL SOURCE FOR DESIGN OF BIOSENSORS. EXPERIMENTAL DEVELOPMENT OF BIOSENSOR BASED ON YfiA GENE Timeline V.P.1*, 9:309:55 9:5510:20 10:2010:45 Coffee/tea break 10:45-11:15 4. 5. 6. 7. 1,2 Rapaport F. , Zinovyev A.1,3*, Barillot E.1, Vert J.-P.2 1 Institute Curie, Bioinformatics Service, Paris, France 2 Ecole des Mines de Paris, Center for Computational Biology, Fontainebleau, France 3 Institute of Computational Modelling SB RAS, Krasnoyarsk, Russia SPECTRAL ANALYSIS OF GENE EXPRESSION PROFILES USING GENE NETWORKS Gunbin К.V.1*, Kogai V.V.2, Fadeev S.I.2, Omelyanchuk L.V.1 1 Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia 2 Sobolev Institute of Mathematics SB RAS, Novosibirsk, Russia A MODEL FOR SENSING THE MORPHOGENE HEDGEHOG CONCENTRATION GRADIENT Galimzyanov A.V. Department of Physicochemical Biology and Epigenetics, Ufa Research Center RAS, Ufa, Russia GENE NETWORKS BEHAVIOR IN A SERIES OF SUCCESIVE CELL DIVISIONS Milanesi L.*, Alfieri R., Merelli I. CNR Institute of Biomedical Technologies, Milan, Italy HCCDB: DATA MINING SYSTEM FOR HUMAN CELL CYCLE GENE 12:55-14:30 Lunch break 16 11:1511:40 11:4012:05 12:0512:30 12:3012:55 July 22, Saturday House of Scientists, Small Conference Hall Morning Session 9:30-12:50 COMPUTATIONAL SYSTEMS BIOLOGY Co-chairpersons: Prof. Ralf Hofestaedt, Bielefeld University, AG Bioinformatics, Bielefeld, Germany Dr. Vyatcheslav Mordvinov, Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia No. 1. 2. 3. Author(s) and Title of Talk Omelianchuk N.A.1*, Mironova V.V.1, Poplavsky A.S.1, Pavlov K.S.1, Savinskaya S.A.2, Podkolodny N.L.1, Mjolsness E.D.3, Meyerowitz E.M.4, Kolchanov N.A.1 1 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia 2 Novosibirsk State University, Novosibirsk, Russia 3 Institute for Genomics and Bioinformatics; University of California, Irvine, USA 4 Division of Biology, California Institute of Technology, Pasadena, USA AGNS (ARABIDOPSIS GENENET SUPPLEMENTARY DATABASE), RELEASE 3.0 Ponomaryov D.1*, Omelianchuk N.2, Mironova V.2, Kolchanov N.2, Mjolsness E.3, Meyerowitz E.4 1 Institute of Informatics Systems, SB RAS, Novosibirsk, Russia 2 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia 3 Institute for Genomics and Bioinformatics, University of California, Irvine, USA 4 Division of Biology, California Institute of Technology, Pasadena, USA A PROGRAM METHOD FOR INFERRING RELATIONSHIPS BETWEEN PHENOTYPIC ABNORMALITIES IN ARABIDOPSIS Akberdin R.*2, Ozonov E.A.1, Mironova V.V.2, Komarov A.V.1, Omelianchuk N.A.2 1 Novosibirsk State University, Novosibirsk, Russia 2 Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia A CELLULAR AUTOMATON TO MODEL THE DEVELOPMENT OF SHOOT MERISTEMS IN ARABIDOPSIS THALIANA Timeline 9:309:55 9:5510:20 10:2010:45 Coffee/tea break 10:45-11:15 4. 5. 6. 7. Nikolaev S.V.1*, Fadeev S.I.2, Penenko A.V.2, Belavskaya V.V.1, Kogay V.V.2, Mjolsness E.3, Kolchanov N.A.1,4 1 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia 2 Institute of Mathematics, SB RAS, Novosibirsk, Russia 3 School of Information and Computer Science, and Institute for Genomics and Bioinformatics, University of California, Irvine, USA 4 Novosibirsk State University, Novosibirsk, Russia CELLULAR-LEVEL MODELLING OF PLANT TISSUE GROWTH AND MORPHOGENESIS Ratushny A.V. Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia IN SILICO CELL: HIERARCHICAL APPROACH AND GENERALIZED HILL FUNCTION METHOD FOR GENETIC SYSTEMS AND METABOLIC PATHWAYS MODELING Rudenko V.M.*, Korotkov E.V. Center of Bioengineering RAS, Moscow, Russia CONSTRUCTION OF THE HCV-HEPATOCYTE SYSTEM MODEL Olsson B.*, Gawronska B., Erlendsson B., Lindlöf A., Dura E. School of Humanities and Informatics, University of Skövde, Sweden AUTOMATED TEXT ANALYSIS OF BIOMEDICAL ABSTRACTS APPLIED TO THE EXTRACTION OF SIGNALING PATHWAYS INVOLVED IN PLANT COLD-ADAPTATION 12:50-14:30 Lunch break 17 11:1511:40 11:4012:05 12:0512:25 12:2512:50 July 22, Saturday House of Scientists, Small Conference Hall Evening Session 14:30-16:40 COMPUTATIONAL SYSTEMS BIOLOGY Co-chairpersons: Dr. Luciano Milanesi, CNR Institute of Biomedical Technologies, Milan, Italy Dr. Mark Lohmann, Cologne University Bioinformatics Center (CUBIC), Cologne, Germany No. 1. 2. Author(s) and Title of Talk 2 E.E.1, 3 Timeline 3 1 Aman Demenkov P.S. , Pintus S.S. , Nemiatov A.I. , Apasieva N.V. , Korotkov R.O. 3, Ignatieva E.V.1, Podkolodny N.L.1, Ivanisenko V.A.*1 1 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia 2 Institute of Mathematics, SB RAS, Novosibirsk, Russia 3 Novosibirsk State University, Novosibirsk, Russia A COMPUTER SYSTEM FOR THE AUTOMATED RECONSTRUCTION OF MOLECULAR-GENETIC INTERACTION NETWORKS Kouznetsova T.N.1*, Ignatieva E.V.1, Oshchepkov D.Yu.1, Kondrakhin Y.V.1, Katokhin A.V.1,2, Shamanina M.Y.1, Mordvinov V.A.1 1 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia 2 Novosibirsk State University, Novosibirsk, Russia A GENE NETWORK OF ADIPOCYTE LIPID METABOLISM: COORDINATED INTERACTIONS BETWEEN THE TRANSCRIPTION FACTORS SREBP, LXR AND PPAR 14:3014:55 14:5515:20 Coffee/tea break 15:20-15:50 3. 4. 2* Bezmaternykh K.D. , Nikulichev Yu.V.1, Likhoshvai V.A.2, Matushkin Yu.G.2, Latipov A.F.1, Kolchanov N.A.2 1 Institute of Theoretical and Applied Mechanics SB RAS, Novosibirsk, Russia 2 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia OPTIMAL CONTROL TASKS IN TERMS OF THE GENE NETWORK MODELS Kolchanov N.A.1*, Orlova G.V.1, Bachinsky A.G.2, Bazhan S.I.2, Shvarts Ya.Sh.3, Golomolzin V.V.1, Popov D.Yu.1, Efimov V.M.5, Tololo I.V.6, Ananko E.A.1, Podkolodnaya O.A.1, Il’ina E.N. 4, Rogov S.I. 4, Tretiakov V.E.4, Kubanova A.A.7, Govorun V.M.4 1 Institute of Cytology and Genetics, SB RAS, Novosibirsk 2 The State Research Center of Virology and Biotechnology “Vector”, Novosibirsk 3 Institute of Internal Medicine SB RAMS, Novosibirsk 4 Institute of Physical and Chemical Medicine MZ RF, Moscow 5 Institute of Animal Taxonomy and Ecology, SB RAS, Novosibirsk 6 Institute of Automation and Electrometry, SB RAS, Novosibirsk 7 Central Research Institute of Dermatovenerology MZ RF, Moscow EPI-GIS: GIS ASSISTED COMPUTER TOOLS FOR DATA ACCUMULATION, COMPUTER ANALYSIS AND MODELING IN MOLECULAR EPIDEMIOLOGY 16:40- 17:20 Closing Ceremony. Summaries from session chairpersons 18 15:5016:15 16:1516:40 July 19, Wednesday House of Scientists, Small Conference Hall Foyer 9:30-12:30 POSTER SESSION Poster Size: The maximum size allowed for each poster is 0.7m wide 1.2m high. Please note that this is a portrait (vertical) poster. Authors must bring their posters printed and ready, as no facilities to produce posters at the Conference are available. Organizers will provide boards and push pins to display your posters. Posters should be readable from a distance of about two to three feet—for easy reading by several people at one time. Presentations may be posted starting from 10:00 on July 17 in the Small Conference Hall Foyer. The posters will be displayed for the overall duration of the Conference. A scheduled Poster Session, only one for all Conference sections, will be held on July 19 from 9:30 to 12:30 in the Small Conference Hall Foyer. During the allotted time, presenters are expected to remain at their individual displays to be available for questions and informal discussion of the poster content. Material should be removed no later than 13.00 on July 22. In the case the author(s) does not take away the presentation, it will be removed and destroyed by the BGRS’2006 Organizing Committee. If you have any questions, please do not hesitate to contact the Organizing Committee in Room no. 200 at the House of Scientists. POSTER PRESENTATIONS (BY SECTIONS) 1. COMPUTATIONAL STRUCTURAL AND FUNCTIONAL GENOMICS AND TRANSCRIPTOMICS Abnizova I.1, Walter K.1, te Boekhorst R.2 , Gilks W.R.1 1 Biostatistics Unit MRC, Institute of Public Health, Robinson Way, Cambridge, UK 2 University of Hertfordshire, College Lane, Hatfield Campus,UK NEW WAY TO OBTAIN A REGULATORY MOTIF REPRESENTATION DUE TO MOTIF ABUNDANCE LEVEL Ananko E.A.*, Kondrakhin Yu.V., Merkulova T.I. Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia PREDICTION OF INTERFERON-INDUCIBLE GENES IN HUMAN GENOME Apasyeva N.V. 1, Yudin N.S.1*, Ignatieva E.V.1, Voevoda M.I.1,2, Romashenko A.G.1 1 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia 2 Institute of Intrnal Medicine, SB RAMS, Novosibirsk, Russia A DATABASE DESIGNED FOR THE POLYMORPHISMS OF THE HUMAN CCR2 GENE 19 Atambaeva S.A., Ivashchenko A.T.*, Khailenko V., Boldina G., Turmagambetova A. al-Farabi Kazakh National University, Almaty, Kazakhstan LENGTH OF EXONS AND INTRONS IN GENES OF SOME HUMAN CHROMOSOMES Atambaeva S.A., Ivashchenko A.T.* al-Farabi Kazakh National University, Almaty, Kazakhstan THE EXONS AND INTRONS LENGTHS IN ARABIDOPSIS THALIANA AND CAENORHABDITIS ELEGANS GENES te Boekhorst R.1*, Walter K.2, Elgar G.3, Gilks W.R.2, Abnizova I.2 1 University of Hertfordshire, College Lane, Hatfield Campus,UK 2 MRC-BSU, Robinson Way, Cambridge, UK 3 Queen’s Mary College, London, UK STATISTICAL CHARACTERIZATION OF CONSERVED NON-CODING ELEMENTS IN VERTEBRATES Califano A.1, Margolin A.1, Fust A.1, Floratos A.1, Badii B.1, Daly E.1, Kim F.1, Ahmady H.1, Nemenman I.1,2, Watkinson J.1, Wang K.1, Smith K.1, Kustagi M.1, Hall M.1, Banerjee N.1, Morozov P.S.1, Hanasoge S.1, Zhang X.1, Wang X.1, Li Z.1 1 Joint Centers for Systems Biology at Columbia University 2 Los Alamos National Laboratory, Los Alamos, USA GEWORKBENCH: A BIOINFORMATIC PLATFORM FOR INTEGRATED GENOMICS Katyshev A.I.1, Rogozin I.B.2, Konstantinov Yu.M.1* 1 Siberian Institute of Plant Physiology and Biochemistry, SB RAS, Irkutsk, Russia 2 Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia IDENTIFICATION OF NEW SUPEROXIDE DISMUTASE TRANSCRIPTS IN PLANTS BY EST ANALYSIS: ALTERNATIVE POLYADENYLATION AND SPLICING EVENTS Kayumov A.R.*, Kirillova J. M., Mikhailova E. O., Balaban N.P., Sharipova M.R. Kazan State University, Kazan, Russia THE PREDICTION OF REGULATION OF SUBTILISIN-LIKE PROTEINASE GENE FROM BACILLUS INTERMEDIUS THROUGH ITS REGULATORY SEQUENCE ANALYSIS Khomicheva I.V.*, Levitsky V.G., Omelianchuk N.A., Ponomarenko M.P. Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia PATTERN OF LOCALLY POSITIONED DINUCLEOTIDES IN microRNA RELATES TO ITS ACCUMULATION LEVELS Khomicheva I.V.1, Levitsky V.G.1, Vishnevsky O.V.1, Savinskaya S.A.2, Omelianchuk N.A.1 1 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia 2 Novosibirsk State University, Novosibirsk, Russia IDENTIFICATION OF ARABIDOPSIS THALIANA microRNAs AMONG MPSS SIGNATURES Khomicheva I.V.*1,2, Vityaev E.E.2, Shipilov T.I.2, Levitsky V.G.1 1 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia 2 Sobolev Institute of Mathematics, SB RAS, Novosibirsk, Russia TRANSCRIPTION FACTOR BINDING SITES RECOGNITION BY THE ExpertDiscovery SYSTEM BASED ON THE RECURSIVE COMPLEX SYGNALS 20 Kolchanov N.A.*, Podkolodnaya O.A., Ananko E.A., Ignatieva E.V., Stepanenko I.L., Khlebodarova T.M., Merkulov V.M., Merkulova T.I., Podkolodny N.L., Romashchenko A.G. Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia TRRD: A DATABASE ON EXPERIMENTALLY IDENTIFIED TRANSCRIPTION REGULATORY REGIONS AND TRANSCRIPTION FACTOR BINDING SITES Kondrakhin Yu.V.*, Ananko E.A., Merkulova T.I. Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia METHODS FOR RECOGNITION OF INTERFERON-INDUCIBLE SITES, PROMOTERS, AND ENHANCERS Lyubetskaya E.V., Seliverstov A.V.*, Lyubetsky V.A. Institute for Information Transmission Problems RAS, Moscow, Russia DETECTING HAIRPINS IN 3’-UNTRANSLATED REGIONS OF HIGHLY EXPRESSED GENES IN ACTINOBACTERIA Merkulov V.M.*, Merkulova T.I. Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia STRUCTURAL VARIANTS OF BINDING SITES FOR GLUCOCORTICOID RECEPTOR AND THE MECHANISMS OF GLUCOCORTICOID REGULATION: ANALYSIS OF GR-TRRD DATABASE Mitra C.K. School of Life Sciences, University of Hyderabad, Hyderabad, India STUDIES ON TRANSCRIPTIONAL REGULATION IN DNA Morozov P.S.1, Morozova I.N.2, Pampou S.2, Shuman H.3, Kalachikov S.M.2 1 Joint Centers for Systems Biology, Columbia University, New York, USA 2 Columbia Genome Center, Columbia University, New York, USA 3 Department of Microbiology, Columbia University, New York, USA UNIQUE OLIGOTIDE BASED MICROARRAY DESIGN FOR THE STUDY OF GENE EXPRESSION OF THE ACCIDENTAL HUMAN PATHOGEN LEGIONELLA PNEUMOPHILA Ogurtsov A.Yu.1*, Vasilchenko A.S.2, Vlasov P.K.3, Shabalina S.A.1, Kondrashov A.S.1, Roytberg M.A.2* 1 National Center for Biotechnology Information, NIH, Bethesda, USA 2 Institute of Mathematical Problems in Biology, Pushchino, Moscow Region, Russia 2 Institute of Molecular Biology, Moscow, Russia OWEN-SCRIPT – AN EXTENDED TOOL FOR PAIRWISE GENOME ALIGNMENT Proskura A.L.1*, Levitsky V.G.1,2, Ignatieva E.V.1,2 1 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia 2 Novosibirsk State University, Novosibirsk, Russia THE ANALYSIS OF SREBP BINDING SITES DISTRIBUTION IN GENE REGIONS BY COMBINED SITEGA AND PWM APPROACH Ryazansky S.S. Institute of Molecular Genetics RAS, Moscow, Russia IDENTIFICATION OF microRNAs ENCODED BY DROSOPHILA TRASPOSABLE ELEMENTS 21 Seliverstov A.V.*, Lyubetsky V.A. Institute for Information Transmission Problems RAS, Moscow, Russia TRANSLATION REGULATION IN CHLOROPLASTS Shagimardanova E.I.*, Shamsutdinov T.R., Chastuchina I.B., Sharipova M.R. Kazan State University, Kazan, Russia ANALYSIS OF REGULATORY REGION OF BACILLUS INTERMEDIUS GLUTAMYL ENDOPEPTIDASE GENE Shavkunov K.S. 1, Masulis I.S. 1, Matushkin Yu.G.2, Ozoline O.N.1* 1 Institute of Cell Biophysics RAS, Pushchino, Moscow region, Russia 2 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia ALTERNATIVE TRANSCRIPTION WITHIN PROCARYOTIC GENES PREDICTED BY PROMOTER-SEARCH SOFTWARE Smetanin D.V.*, Chumak N.M. Krasnodar Research Institute of Agriculture, Krasnodar, Russia A STEP BEYOND PLANT TRANSCRIPT`S POLYADENYLATION SITE Smirnova O.G.*, Ibragimova S.S., Grigorovich D.A., Kochetov A.V. Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia TGP (TransGene Promoters): A DATABASE OF BIOTECHNOLOGICALLY IMPORTANT PLANT GENE PROMOTERS Stavrovskaya E.D.1,2, *, Makeev V.J.3, Merkeev I.V.3, Mironov A.A.1,2,3 1 Institute for Information Transmission Problems RAS, Moscow, Russia 2 Department of Bioengineering and Bioinformatics, Moscow State University, Moscow, Russia 3 State Scientific Center GosNIIGenetica, Moscow, Russia TOOL FOR AUTOMATIC DETECTION OF CO-REGULATED GENES Taraskina A.S. 1,3, Cheremushkin E.S.2,3 1 Mechanics and Mathematics Department, Novosibirsk State University, Novosibirsk, Russia 2 Biorainbow Group, Novosibirsk, Russia 3 Ershov Institute of Informatics Systems, Novosibirsk, Russia THE MODIFIED FUZZY C- MEANS METHOD FOR CLUSTERING OF MICROARRAY DATA Titov I.I.1,2 1 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia 2 Novosibirsk State University, Russia THE GArna TOOLBOX FOR RNA STRUCTURE ANALYSIS: THE 2006 STATE OF THE ART Titov I.I.1,2*, Ivanisenko A.Yu.2 1 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia 2 Novosibirsk State University, Novosibirsk, Russia A MODEL OF THE TRANSLATIONAL INHIBITION BY MIRISC COMPLEX DESCRIBES PROTEIN SYNTHESIS VARIATIONS INDUCED BY MUTATIONS IN MAMMALIAN miRNA SITES 22 Vladimirov N.V. *, Likhoshvai V.A., Matushkin Yu.G. Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia ROLES OF CODON BIASES AND POTENTIAL SECONDARY STRUCTURES IN mRNA TRANSLATION OF UNICELLULAR ORGANISMS Vladimirov N.V. *, Kochetov A.V., Grigorovich D.A., Matushkin Yu.G. Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia RSCU_COMPARER: A NEW STATISTICAL TOOL FOR PRACTICAL ANALYSIS OF CODON USAGE Vityaev Е.E.*1,2, Lapardin K.A.2, Khomicheva I.V.3, Levitsky V.G.2,3 1 Institute of Mathematics, SB RAS, Novosibirsk, Russia 2 Novosibirsk State University, Novosibirsk, Russia 3 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia TRANSCRIPTION FACTOR BINDING SITES RECOGNITION BY THE REGULARITIES MATRICES BASED ON THE NATURAL CLASSIFICATION METHOD Volod’ko V.B.1, Shuvaev R.Yu.1, Ulyashin A.V.1, Novikova O.S.2, Matushkin Yu.G.2,3 1 Novosibirsk Center of Information Technologies "UniPro", Novosibirsk, Russia 2 Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia 3 Novosibirsk State University, Novosibirsk, Russia GENOMEBROWSER – SOFTWARE FOR VISUAL SEQUENCE ANALYSIS Zagoruiko N.G. 1*, Kutnenko O.A. 1, Borisova I.A. 1, Kiselev A.N. 1, Ptitsyn A.A.2 1 Institute of Mathematics, SB RAS, Novosibirsk, Russia 2 Pennington Biomedical Research Center, Baton Rouge LA 70808, USA SELECTION OF INFORMATIVE SUBSET OF GENE EXPRESSION PROFILES IN PROGNOSTIC ANALYSIS OF TYPE II DIABETES 2. COMPUTATIONAL STRUCTURAL AND FUNCTIONAL PROTEOMICS Afonnikov D.A.1,2, Morozov A.V.1 1 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia 2 Novosibirsk State University, Novosibirsk, Russia CONNP: THE PREDICTION OF THE CONTACT NUMBERS OF THE AMINO ACID RESIDUES IN PROTEINS USING NEURAL NETWORK REGRESSION Aman E.E. *, Demenkov P.S., Ivanisenko V.A. Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia ANALYSIS OF THE TERTIARY STRUCTURE OF THE PPAR AND RXR TRANSCRIPTIONAL FACTORS AND THEIR MUTANT VARIANTS Bakulina A.Yu.*1, Sineva E.V.2, Solonin A.S.2, Maksyutov A.Z.1 1 State Research Center of Virology and Biotechnology “Vector”, Koltsovo, Novosibirsk region, Russia 2 Institute of Biochemistry and Physiology of Microorganisms, Russian Academy of Sciences, Pushchino, Moscow region, Russia MOLECULAR MODELING OF B. CEREUS HEMOLYSIN II, A PORE-FORMING PROTEIN 23 Baryshev P.B.1*, Afonnikov D.A.1,2, Nikolaev S.V.2 1 Novosibirsk State University, Novosibirsk, Russia 2 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia IDENTIFICATION AND STRUCTURE-FUNCTIONAL ANALYSIS OF THE SPECIFICITY DETERMINING RESIDUES OF THE ALPHA SUBUNITS OF THE PROTEOSOMAL COMPLEX Batsianovsy A.V.*, Vlasov P.K. Engelhard Institute of Molecular Biology, RAS, Moscow, Russia *Moscow State University, Moscow, Russia CLUSTERING ANALYSIS OF CONFORMATIONAL STRUCTURES OF SHORT OLIGOPEPTIDES Chugunov A.O.1,2,*, Novoseletsky V.N.1,3, Efremov R.G.1 1 Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, Moscow, Russia 2 Department of Bioengineering, Biological Faculty, M.V. Lomonosov Moscow State University, Moscow, Russia 3 Moscow Institute of Physics and Technology (State University), Moscow region, Russia A METHOD TO ASSESS CORRECT/MISFOLDED STRUCTURES OF TRANSMEMBRANE DOMAINS OF MEMBRANE PROTEINS Efimov A.V., Brazhnikov E.V., Kondratova M.S. Institute of Protein Research, Russian Academy of Sciences, Pushchino, Moscow region, Russia EFFECT OF THE STRUCTURAL CONTEXT ON SPECIFICITY OF INTRAAND INTERHELICAL INTERACTIONS IN PROTEINS Dulko V., Feranchuk S. United Institute of Informatics Problems of the National Academy of Sciences of Belarus, Minsk, Belarus DYNAMIC PROGRAMMING ALGORITHM PARALLIZATION FOR PROTEIN FOLDING Gou Z., Hwang S., Kuznetsov I.B.* Gen*NY*sis Center for Excellence in Cancer Genomics, University at Albany, Rensselaer, USA SEQUENCE-BASED PREDICTION OF DNA-BINDING SITES ON DNA-BINDING PROTEINS Ivanisenko V.A.1,2*, Ivanisenko T.V.3, Sharonova I.V.2, Krestyanova M.A.2, Ivanisenko N.V.2, Grigorovich D.A.1 1 Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia 2 Novosibirsk State University, Novosibirsk, Russia 3 Siberian State University of Telecommunications and Information Sciences, Novosibirsk, Russia PDBSite DATABASE AND PDBSiteScan TOOL: RECOGNITION OF FUNCTIONAL SITES IN PROTEIN 3d STRUCTURE AND TEMPLATE-BASED DOCKING Konshina A.G.*, Dubinnyi M.A., Efremov R.G. Shemyakin–Ovchinnikov Institute of Bioorganic Chemistry, RAS, Moscow, Russia STRUCTURAL DETERMINANTS OF CARDIOTOXINS’ MEMBRANE BINDING: A MOLECULAR MODELING APPROACH 24 Nizolenko L.Ph.1*, Bachinsky A.G. 1, Yarygin A.A.1, Naumochkin A.N.1, Grigorovich D.A.2 1 SRC VB “Vector”, Koltsovo, Novosibirsk, Russia 2 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia PROF_PAT: THE UPDATED DATABASE OF PROTEIN FAMILY PATTERNS. CURRENT STATUS Nyporko A.Yu.*, Blume Ya.B. Institute of Cell Biology and Genetic Engineering of NAS of Ukraine, Kiev, Ukraine THE FEATURES OF STRUCTURAL DYNAMICS OF DIFFERENT TUBULIN SUBUNITS Polyansky A.A.*1,2, Aliper E.T.1,2, Volynsky P.E.1, Efremov R.G.1 1 M.M. Shemyakin and Yu.A. Ovchinnikov Institute of Bioorganic Chemistry Russian Academy of Sciences, Moscow, Russia 2 Biological Department, M.V.Lomonosov Moscow State University, Moscow, Russia ACTION OF MEMBRANE-ACTIVE PEPTIDES ON EXPLICIT LIPID BILAYERS. ROLE OF SPECIFIC PEPTIDE-LIPID INTERACTIONS IN MEMBRANE DESTABILIZATION Pyrkov T.V.*1,2, Kosinsky Yu.A.1, Arseniev A.S.1, Priestle J.P.3, Jacoby E.3, Efremov R.G.1 1 M.M. Shemyakin and Yu. A. Ovchinnikov Institute of Bioorganic Chemistry RAS, Moscow, Russia 2 Moscow Institute of Physics and Technology, Moscow, Russia 3 Novartis Institutes for Biomedical Research, Basel, Swizerland COMBINING MOLECULAR DOCKING WITH RECEPTOR DOMAIN MOTIONS: SIMULATIONS OF BINDING OF ATP TO CA-ATPASE Shaytan A.K.*, Khokhlov A.R., Ivanov V.A. Chair of Polymer and Crystal Physics, Physics Department, Moscow State University, Moscow, Russia PEPDITE DYNAMICS AT WATER MEMBRANE INTERFACE Slyadnikov E.E. Informatization Problems Department of Tomsk Scientific Centre, SB RAS, Tomsk, Russia MICROSCOPIC MODEL OF CONFORMATION DEGREES OF FREEDOM CYTOSKELETON MICROTUBULE AND ITS STRUCTURAL ANALOG IN OPTICS Troshin P.V.1*, Papiz M.Z.1, Prince S.M.2, Daniel E.J.3, Morris C.3*, Griffiths S.L.4, Diprose J.M.5, Pilicheva K.5, Niekerk J.V.6, Pajon A.7 1* Membrane Protein Structure Initiative, CCLRC Daresbury Laboratory, UK 2 Faculty of Life Sciences, University of Manchester, Sackville St., Manchester, UK 3 Protein information Management System, CCLRC Daresbury Laboratory, UK 4 York Structural Biology Laboratory, University of York, UK 5 Oxford Protein Production Facility, Division of Structural Biology, University of Oxford, UK 6 Scottish Structural Proteomics Facility, University of Dundee, UK 7 European Bioinformatics Institute, Hinxton, UK LABORATORY INFORMATION MANAGEMENT SYSTEM FOR MEMBRANE PROTEIN STRUCTURE INITIATIVE Volkova O.A.1, Kochetov A.V.1* 1 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia AMINO ACID PREFERENCES AT THE N-TERMINAL PART OF EUKARYOTIC PROTEINS CORRELATING WITH A SPECIFIC CONTEXTUAL ORGANIZATION OF TRANSLATION INITIATION SIGNAL 25 3. COMPARATIVE AND EVOLUTIONARY GENOMICS AND PROTEOMICS Babkin I.V.*, Shchelkunov S.N. State Research Center of Virology and Biotechnology “Vector”, Koltsovo, Novosibirsk region, Russia TIME SCALE OF POXVIRUS EVOLUTION Boeva V.A.*1, Makeev V.J.2 1 Moscow State University, Department of Bioengineering and Bioinformatics, Moscow, Russia 2 GosNIIGenetika State Research Center, Moscow, Russia MICRO- AND MINISATELLITES IN HUMAN GENOME, TandemSWAN SOFTWARE IN USE Brinza D., Perelygin A., Brinton M., Zelikovsky A.* Georgia State University, Atlanta, USA SEARCH FOR MULTI-SNP DISEASE ASSOCIATION Bukin Y.S.*, Pudovkina T.A. Limnological Institute of SB RAS, Irkutsk, Russia THE MODELS OF POPULATION DYNAMICS AS TOOL FOR STUDYING OF GENETIC POLYMORPHISM OF BAIKALIAN POLYCHAETS Chumakov M.I., Mazilov S., Zotova T.V. Institute of Symbiology RAS, Saratov, Russia SEARCHING FOR NUCLEOTIDE SEQUENCES LIKE AGROBACTERIAL T-DNA RIGHT BORDER IN PLANT GENOMES Golovnina K.*1, Blinov A.1, Chang L.-S.2 1 Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia 2 Center for Childhood Cancer, Children’s Research Institute, Children’s Hospital and Department of Pediatrics, The Ohio State University, USA EVOLUTION AND ORIGIN OF NEUROFIBROMIN, THE PRODUCT OF THE NEUROFIBROMATOSIS TYPE 1 (NF1) TUMOR-SUPRESSOR GENE Gorbunov K.Yu.*, Lyubetsky V.A. Institute for Information Transmission Problems of the Russian Academy of Sciences, Moscow, Russia INFERRING REGULATIORY SIGNAL PROFILES AND EVOLUTIONARY EVENTS Gunbin K.V., Morozov A.V., Afonnikov D.A. Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia A METHOD FOR SEMIAUTOMATED ANALYSIS OF GENE EVOLUTION Guryev V.1*, Smits B.M.G.1, van de Belt J. 1, Verheul M.1, Hubner N.2 , Cuppen E.1 1 Hubrecht Laboratorium, Utrecht, The Netherlands 2 Max-Delbruck-Center for Molecular Medicine (MDC), Berlin-Buch, Germany HAPLOTYPE BLOCK STRUCTURE IS CONSERVED ACROSS MAMMALS Kabanova A1., Novikova O1., Gunbin K1., Fet V2., Blinov A.1* 1 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia 2 Marshall University, Huntington, WV, USA EVOLUTIONARY RELATIONSHIPS AND DISTRIBUTION OF THE DIFFERENT LTR RETROTRANSPOSON FAMILIES IN PLANTS 26 Khan M. 2, Jamil K.1, 2 * 1 Indo American Cancer Institute and Research Centre, Hyderabad, India 2 School of Biotechnology, (MGNIRSA,) University of Mysore, Mysore, India THE PHYLOGENETIC ANALYSIS OF UBIQUITIN CONJUGATING ENZYMES OF CANCER METASIGNATURE Korostishevsky M.1*, Bonne’-Tamir B.1, Bentwich Z.2, Tsimanis A.2 1 Department of Human Molecular Genetics and Biochemistry, Tel Aviv University, Israel 2 Institute of Clinical Immunology and AIDS, Kaplan Medical Center, Rehovot, Israel MULTI-SNP ANALYSIS OF CCR5-CCR2 GENES IN ETHIOPIAN JEWS: MICRO-EVOLUTION AND HIV-RESISTANCE IMPLICATIONS Kovaleva G.Yu.*1,2, Gelfand M.S.1,2 1 Moscow State University, Department of Bioengeneering and Bioinformatics, Moscow, Russia 2 Institute for Information Transmission Problems RAS, Moscow, Russia TRANSCRIPTIONAL REGULATION OF THE METHIONINE BIOSYNTHESIS IN ACTINOBACTERIA AND STREPTOCOCCI Kuznetsova A.Y.*1, Naumoff D.G.2 1 Moscow State University, Department of Bioengineering and Bioinformatics, Moscow, Russia 2 State Institute for Genetics and Selection of Industrial Microorganisms, Moscow, Russia PHYLOGENETIC ANALYSIS OF COG1649, A NEW FAMILY OF PREDICTED GLYCOSYL HYDROLASES Lashin S.A.*, Likhoshvai V.A., Kolchanov N.A, Matushkin Yu.G. Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia EVOLUTIONARY CONSTRUCTOR: A PACKAGE FOR MODELING COEVOLUTION OF UNICELLULAR ORGANISMS Lashin S.A.*, Likhoshvai V.A., Kolchanov N.A., Matushkin Yu.G. Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia MODELING OF HORIZONTAL GENE TRANSFER IN PROKARYOTIC POPULATIONS WITH THE “EVOLUTIONARY CONSTRUCTOR” PROGRAM PACKAGE Liventseva V.*, Kaygorodova I. Limnological Institute, SB RAS, Irkutsk, Russia MOLECULES VERSUS MORPHOLOGY IN OLIGOCHAETA SYSTEMATICS Novikova O.*1, Fursov M.2, Blinov A.1 1 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia 2 Novosibirsk Center of Information Technologies “UniPro”, Novosibirsk, Russia NEW FAMILY OF LTR RETOTRANSPOSABLE ELEMENTS FROM FUNGI Peretolchina T.E.*, Bukin Yu.S., Sitnikova T.Ya., Sherbakov D.Yu. Limnological Institute, SB RAS, Irkutsk, Russia POPULATION GENETIC POLYMORPHISM OF ENDEMIC MOLLUSCS BAICALIA CARINATA (MOLLUSCA: CAENOGASTROPODA) 27 Rotskaya U.N., Rogozin I.B., Vasyunina E.A., Kolosova N.G., Sinitsyna O.I.* Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia THEORETICAL ANALYSIS OF THE MITOCHONDRIAL DNA SOMATIC MUTATION SPECTRA IN OXYS AND WISTAR RATS Rusin L.Y.*, Lyubetsky V.A. Institute for Information Transmission Problems RAS, Moscow, Russia REFINEMENT OF PHYLOGENETIC SIGNAL IN MULTIPLE SEQUENCE ALIGNMENT: RESULTS OF SIMULATION STUDY Ryabinina O.M. Vavilov Institute of General Genetics RAS, Moscow, Russia GENETIC DIVERSITY AND PHYLOGENETIC RELATIONSHIPS IN GROUPS OF ASIAN GUARDIAN, SIBERIAN HUNTING AND EUROPEAN SHEPHERD DOG BREEDS Yudin N.S.1*, Vasil’eva L.A.1, Kobzev V.F.1, Kuznetsova T.N.1, Ignatieva E.V.1, Oshchepkov D.Yu.1, Voevoda M.I.1,2, Romaschenko A.G.1 1 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia 2 Institute of Internal Medicine, SB RAMS, Novosibirsk, Russia ASSOCIATION STUDY OF SNP OF THE TNF-ALPHA GENE WITH BOVINE LEUKOSIS AND EVALUATION OF ITS FUNCTIONAL SIGNIFICANCE Zotov V.S.1, Punina N.V.1, Dorokhov D.B.1, Schaad N.W.2, Ignatov A.N.1* 1 Centre “Bioengineering” RAS, Moscow, Russia 2 FDWSRU, USDA-ARS, MD, USA PHYLOGENETIC CHANGES IN CHLOROPLAST GENOMES 4. COMPUTATIONAL SYSTEMS BIOLOGY 4.1 Modeling of Molecular Genetic Systems and Processes in Bacterial Cell Khlebodarova T.M.1, Ananko E.A.1*, Nedosekina E.A.1, Podkolodnaya O.A.1, Poplavsky A.S.1, Smirnova O.G.1, Ibragimova S.S.1, Mishchenko E.L.1, Stepanenko I.L.1, Ignatieva E.V.1, Proskura A.L.1, Nishio Y.2, Imaizumi A.2, Usuda Y.2, Matsui K.2, Podkolodny N.L.1, Kolchanov N.A.1 1 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia 2 Institute of Life Sciences, Ajinomoto, Co., Inc., Kawasaki-city, Japan IN SILICO CELL II. INFORMATION SOURCES FOR MODELING ESCHERICHIA COLI METABOLIC PATHWAYS Ratushny A.V.1*, Usuda Y.2, Matsui K.2, Podkolodnaya O.A.1 1 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia 2 Institute of Life Sciences, Ajinomoto, Co., Inc., Kawasaki, Japan MATHEMATICAL MODELING OF ELEMENTARY PROCESSES OF THE GENE NETWORK CONTROLLING HISTIDINE BIOSYNTHESIS IN ESCHERICHIA COLI Nedosekina E.A.1*, Usuda Y.2, Matsui K.2 1 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia 2 Institute of Life Sciences, Ajinomoto, Co., Inc., Kawasaki, Japan MODELING OF THE EFFECTS OF THREONINE, VALINE, ISOLEUCINE AND PYRIDOXAL 5'-MONOPHOSPHATE ON BIOSYNTHETIC THREONINE DEHYDRATASE REACTION 28 Ratushny A.V.*, Nedosekina E.A. Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia REGULATION OF PYRIMIDINE BIOSYNTHESIS IN ESCHERICHIA COLI: GENE NETWORK RECONSTRUCTION AND MATHEMATICAL MODELING Ratushny A.V.1*, Smirnova O.G.1, Usuda Y.2, Matsui K.2 1 Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia 2 Institute of Life Sciences, Ajinomoto, Co., Inc., Kawasaki, Japan REGULATION OF THE PENTOSE PHOSPHATE PATHWAY IN ESCHERICHIA COLI: GENE NETWORK RECONSTRUCTION AND MATHEMATICAL MODELING OF METABOLIC REACTIONS Ananko E.A.1*, Ratushny A.V. 1, Usuda Y.2, Matsui K.2 1 Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia 2 Institute of Life Sciences, Ajinomoto, Co., Inc., Kawasaki, Japan AROMATIC AMINO ACID BIOSYNTHESIS IN ESCHERICHIA COLI: GENERALIZED HILL FUNCTION MODEL OF THE TRYPTOPHAN-SENSITIVE 3-DEOXY-D-ARABINOHEPTULOSONATE-7-PHOSPHATE SYNTHASE REACTION DEMONSTRATE COMPLICATED MECHANISM Ratushny A.V.*, Khlebodarova T.M. Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia MATHEMATICAL MODELING OF REGULATION OF cyоABCDE OPERON EXPRESSION IN ESCHERICHIA COLI Khlebodarova T.M.*, Lashin S.A., Apasieva N.V. Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia GENE NETWORK RECONSTRUCTION AND MATHEMATICAL MODELING OF ESCHERICHIA COLI RESPIRATION: REGULATION OF F0F1-ATP SYNTHASE BY METAL IONS Nedosekina E.A. Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia MATHEMATICAL MODELING OF REGULATION OF ESCHERICHIA COLI PURINE BIOSYNTHESIS PATHWAY ENZYMATIC REACTIONS Mishchenko E.L. 1*, Lashin S.A.1, Usuda Y.2, Matsui K.2 1 Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia 2 Institute of Life Sciences, Ajinomoto, Co., Inc., Kawasaki, Japan RECONSTRUCTION AND MATHEMATICAL MODELING OF THE GENE NETWORK CONTROLLING CYSTEINE BIOSYNTHESIS IN ESCHERICHIA COLI: REGULATION OF SERINE ACETYLTRANSFERASE ACTIVITY Smirnova O.G.*, Ratushny A.V. Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia GENE NETWORK RECONSTRUCTION AND MATHEMATICAL MODELING OF SALVAGE PATHWAYS: REGULATION OF ADENINE PHOSPHORIBOSYLTRANSFERASE ACTIVITY BY STRUCTURALLY SIMILAR SUBSTRATES 29 Turnaev I.I.1*, Ibragimova S.S.1, Usuda Y.2, Matsui K.2 1 Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia 2 Institute of Life Sciences, Ajinomoto, Co., Inc., Kawasaki, Japan MATHEMATICAL MODELING OF SERINE AND GLYCINE BIOSYNTHESIS REGULATION IN ESCHERICHIA COLI Turnaev I.I.*, Smirnova O.G. Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia REGULATION OF PYRUVATE BIOSYNTHESIS IN ESCHERICHIA COLI: GENE NETWORK RECONSTRUCTION AND MATHEMATICAL MODELING OF ENZYMATIC REACTIONS OF THE PATHWAY Ibragimova S.S.*, Smirnova O.G., Grigorovich D.A., Khlebodarova T.M. Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia BiotechPro: A DATABASE FOR MICROBIOLOGICALLY SYNTHESIZED PRODUCTS OF BIOTECHNOLOGICAL VALUE Peshkov I.M.1, Fadeev S.I.2 1 Novosibirsk State University, Novosibirsk, Russia 2 Sobolev Institute of Mathematics SB RAS, Novosibirsk, Russia ABOUT RECONSTRUCTION OF REGULATORY MECHANISM OF GENE ELEMENTS’ EXPRESSION 4.2 Modeling of Molecular Genetic Systems and Processes in Multicellular Organisms Akberdin I.R.1*, Kashevarova N.A.2, Khlebodarova T.M.1, Bazhan S.I.3, Likhoshvai V.A.1 1 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia 2 Novosibirsk State University, Novosibirsk, Russia 3 State Research Center of Virology and Biotechnology “Vector”, Koltsovo, Russia A MATHEMATICAL MODEL FOR THE INFLUENZA VIRUS LIFE CYCLE Bazhan S.I.1*, Schwartz Ya.Sh.2, Gainova I.A.3, Ananko E.A.4 1 State Research Center of Virology and Biotechnology “Vector”, Koltsovo, Russia 2 Institute of Internal Medicine, SB RAMS, Novosibirsk 3 Institute of Mathematics, SB RAS, Novosibirsk 4 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia A MATHEMATICAL MODEL OF IMMUNE RESPONSE IN INFECTION INDUCED BY MYCOBACTERIA TUBERCULOSIS. PREDICTION OF THE DISEASE COURSE AND OUTCOMES AT DIFFERENT TREATMENT REGIMENS Bezmaternikh K.D., Mishchenko E.L.*, Ratushny A.V., Likhoshvai V.A., Khlebodarova T.M., Ivanisenko V.A. Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia A MINIMUM MATHEMATICAL MODEL FOR SUPPRESSION HCV RNA REPLICATION IN CELL CULTURE Fadeev S.I.1*, Shtokalo D.N.2, Likhosvai V.A.3 1 Sobolev Institute of Mathematics SB RAS, Novosibirsk, Russia 2 Novosibirsk State University, Novosibirsk, Russia 3 Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia MATRIX PROCESS MODELLING: STUDY OF A MODEL OF SYNTHESIS OF LINEAR BIOMOLECULES WITH REGARD TO REVERSIBILITY OF PROCESSES 30 Gursky V.V.1*, Samsonov A.M.1, Reinitz J.2 1 Ioffe Physico-Technical Institute of the Russian Academy of Sciences, St. Petersburg, Russia 2 Stony Brook University, Stony Brook, New York, USA APPROXIMATE STATIONARY ATTRACTORS IN DROSOPHILA GAP GENE CIRCUITS IN THE LIMIT OF STEEP-SIGMOID INTERACTIONS Kalashnikova E.V.1, Dymshits G. M.1, Kolpakov F.A.2,3 1 Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia 2 Design Technological Institute of Digital Techniques SB RAS, Novosibirsk, Russia 3 Institute of Systems Biology OOO, Novosibirsk, Russia SIGNAL TRANSDUCTION PATHWAYS INVOLVED IN TRANSCRIPTIONAL REGULATION OF TYROSINE HYDROXYLASE Mischenko E.L.1, Korotkov R.O.2, Ivanisenko V.A.1,2* 1 Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia 2 Novosibirsk State University, Novosibirsk, Russia HCV-KINET DATABASE: KINETIC PARAMETER REACTIONS AND REGULATORY PROCESSES OF THE LIFE CYCLE OF THE HEPATITIS C VIRUS Ratushny A.V. Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia MATHEMATICAL MODELING OF RECEPTOR MEDIATED ENDOCYTOSIS OF LOW-DENSITY LIPOPROTEINS AND THEIR DEGRADATION IN LYSOSOMES Ratushny A.V. Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia MATHEMATICAL MODELING OF THE GENE NETWORK CONTROLLING HOMEOSTASIS OF INTRACELLULAR CHOLESTEROL Ratushny A.V.*, Bezmaternikh K.D Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia CONSERVED PROPERTIES OF ENZYMATIC SYSTEMS: PRENYLTRANSFERASE KINETICS Sharipov R.N.1,2,3,*, Kolpakova A.F.4 1 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia 2 Design Technological Institute of Digital Techniques SB RAS, Novosibirsk, Russia 3 Institute of Systems Biology OOO, Novosibirsk, Russia 4 State Research Institute for Medical Problems of the North, SB RAMS, Krasnoyarsk, Russia FORMAL DESCRIPTION OF NF-B PATHWAY, ITS ROLE IN INFLAMMATION, INHIBITION OF APOPTOSIS, CARCINOGENESIS, AND WAYS OF INACTIVATION FOR PREDICTION OF NEW TARGETS FOR ANTI-INFLAMMATORY AND ANTI-CANCER TREATMENT Stepanenko I.L1*., Podkolodnaya N.N.1, Paramonova N.V.1, Podkolodny N.L.1,2 1 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia 2 Institute of Computational Mathematics and Mathematical Geophysics SB RAS, Novosibirsk, Russia RECONSTRUCTION AND STRUCTURE ANALYSIS OF APOPTOSIS GENE NETWORK IN HEPATITIS C 31 4.3 Modeling of morphogenesis Bukharina T.A.1, Katokhin A.V.1,2, Furman D.P.1,2 1 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia 2 Novosibirsk State University, Novosibirsk, Russia THE GENE NETWORK DETERMINING DEVELOPMENT OF DROSOPHILA MELANOGASTER MECHANORECEPTORS Mironova V.V.1*, Poplavsky A.S.1, Ponomarev D.K.2, Omelianchuk N.A.1 1 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia 2 Institute of Informatics System, tait SB RAS, Novosibirsk, Russia ONTOLOGY OF ARABIDOPSIS GENE NET SUPPLEMENTARY DATABASE (AGNS), CROSS DATABASE REFERENCES TO TAIR ONTOLOGY Nikolaev S.V.1*, Fadeev S.I.2, Kogay V.V.2, Mjolsness E.3, Kolchanov N.A.1 1 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia 2 Institute of Mathematics SB RAS, Novosibirsk, Russia 3 School of Information and Computer Science, and Institute for Genomics and Bioinformatics, University of California, Irvine, USA A ONE-DIMENSIONAL MODEL FOR THE REGULATION OF THE SIZE OF THE RENEWABLE ZONE IN BIOLOGICAL TISSUE Nikolaev S.V.1*, Penenko A.V.2, Belavskaya V.V.1, Mjolsness E.3, Kolchanov N.A.1 1 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia 2 Institute of Mathematics, SB RAS, Novosibirsk, Russia 3 School of Information and Computer Science, and Institute for Genomics and Bioinformatics, University of California, Irvine, USA A SYSTEM FOR SIMULATION OF 2D PLANT TISSUE GROWTH AND DEVELOPMENT Ponomaryov D.1*, Omelianchuk N.2, Kolchanov N.2, Mjolsness E.3, Meyerowitz E.4 1 Institute of Informatics Systems, SB RAS, Novosibirsk, Russia 2 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia 3 Institute for Genomics and Bioinformatics, University of California, Irvine, USA 4 Division of Biology, California Institute of Technology, Pasadena, CA, USA SEMANTICALLY RICH ONTOLOGY OF ANATOMICAL STRUCTURE AND DEVELOPMENT FOR ARABIDOPSIS THALIANA L. 4.4 Gene networks theory: mathematical problems and software Aman E.E.*, Levitsky V.G., Ignatieva E.V. Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia RECONSTRUCTION AND COMPUTER ANALYSIS OF THE GENE NETWORK OF FATTY ACID -OXIDATION REGULATED BY THE PPAR TRANSCRIPTIONAL FACTORS Demidenko G.V.1*, Khropova Yu.E.2 1 Sobolev Institute of Mathematics, SB RAS, Novosibirsk, Russia 2 Novosibirsk State University, Novosibirsk, Russia MATRIX PROCESS MODELLING: ON PROPERTIES OF SOLUTIONS OF ONE DELAY DIFFERENTIAL EQUATIONS 32 Demidenko G.V.1*, Khropova Yu.E.2, Kotova T.V.2 1 Sobolev Institute of Mathematics, SB RAS, Novosibirsk, Russia 2 Novosibirsk State University, Novosibirsk, Russia MATRIX PROCESS MODELLING: ON ONE CLASS OF INFINITE-ORDER SYSTEMS OF DIFFERENTIAL EQUATIONS AND ON DELAY DIFFERENTIAL EQUATIONS Demidenko G.V.1*, Mudrov A.V. 2 1 Sobolev Institute of Mathematics, SB RAS, Novosibirsk, Russia 2 Novosibirsk State University, Novosibirsk, Russia MATRIX PROCESS MODELLING: ON A NEW METHOD OF APPROXIMATION OF SOLUTIONS OF DELAY DIFFERENTIAL EQUATIONS Fadeev S.I.*, Korolev V.K. Sobolev Institute of Mathematics, SB RAS, Novosibirsk, Russia PROGRAM PACKAGE HGNET FOR COMPUTATIONAL INVESTIGATION OF HYPOTHETICAL GENE NETWORKS Klishevich M.A.*, Kogai V.V., Fadeev S.I. Sobolev Institute of Mathematics, SB RAS, Novosibirsk, Russia RESEARCH OF CYCLIC GENE NETWORK CIRCUITS WITH NEGATIVE TYPE OF REGULATION Kolpakov F.1,2*, Sharipov R.3,1,2, Cheremushkina E.2, Kalashnikova E.3 1 Institute of Systems Biology OOO, Novosibirsk, Russia 2 Design Technological Institute of Digital Techniques, SB RAS, Novosibirsk, Russia 3 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia BIOPATH – A NEW APPROACH TO FORMALIZED DESCRIPTION AND SIMULATION OF BIOLOGICAL SYSTEMS Komarov A.V.1*, Akberdin I. R.2, Ozonov E. A.1, Evdokimov A. A.3 1 Novosibirsk State University, Novosibirsk, Russia 2 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia 3 Sobolev Institute of Mathematics, SB RAS, Novosibirsk, Russia ON THE RECONSTRUCTION OF GENETIC AUTOMATE ON THE BASIS OF BOOLEAN DYNAMIC DATA Likhoshvai V.A.1*, Rudneva D.S.2, Fadeev S.I.3 1 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia 2 Novosibirsk State University, Novosibirsk, Russia 3 Sobolev Institute of Mathematics, SB RAS, Novosibirsk, Russia OSCILLATIONS OF CHAOTIC TYPE IN SYMMETRIC GENE NETWORKS OF SMALL DIMENSION Matveeva I.I. 1, Popov A.M. 1 Sobolev Institute of Mathematics, SB RAS, Novosibirsk, Russia 2 Novosibirsk State University, Novosibirsk, Russia MATRIX PROCESS MODELLING: DEPENDENCE OF SOLUTIONS OF A SYSTEM OF DIFFERENTIAL EQUATIONS ON PARAMETER 33 Podkolodny N.L.1,2*, Podkolodnaya N.N.1, Miginsky D.S.1, Poplavsky A.S.1, Likhoshvai V.A.1, Compani B.3, Mjolsness E.3 1 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia 2 Institute of Computational Mathematics and Mathematical Geophysics, SB RAS, Novosibirsk, Russia 3 Institute for Genomics and Bioinformatics, University of California, Irvine, USA AN INTEGRATION OF THE DESCRIPTIONS OF GENE NETWORKS AND THEIR MODELS PRESENTED IN SIGMOID (CELLERATOR) AND GENENET Tchuraev R.N. Department of Physicochemical Biology and Epigenetics, Ufa Research Center, RAS, Ufa, Russia RESEARCH ON BEHAVIOR OF GOVERNING GENE/EPIGENE NETWORKS AS A PROBLEM OF CELLULAR AUTOMATA IDENTIFICATION 5. NEW APPROACHES TO ANALYSIS/MODELING OF BIOMOLECULAR DATA AND PROCESSES Arrigo P.1*, Cardo P.P.2 1 CNR Institute for Macromolecular Studies, Genoa, Italy 2 Department of Health Science, University of Genoa, Genoa, Italy INVESTIGATION OF NAMED ENTITY RECOGNITION IN MOLECULAR BIOLOGY BY DATA FUSION Axenovich M.A. Department of Mathematics, Iowa State University, Ames, USA EDIT DISTANCE IN COMBINATORIAL STRUCTURES Axenovich T.I. Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia GRAPH THEORY ALGORITHM FOR SOLUTION OF COMPUTATIONAL PROBLEMS OF GENE MAPPING Huang Shaoqing *, Cui Yini College of Life Science and Biotechnology, Shanghai Jiao Tong University, Shanghai, China UNEQUALLY SPACED SAMPLING MAPPING OF QTL Kolpakov F.1,2,*, Poroikov V.3, Sharipov R.4,2,1, Milanesi L.5, Kel A.6 1 Design Technological Institute of Digital Techniques, SB RAS, Novosibirsk, Russia 2 Institute of Systems Biology, Novosibirsk OOO, Russia 3 Institute of Biomedical Chemistry, RAMS, Moscow, Russia 4 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia 5 Institute of Biomedical Technologies, Segrate (MI), Italy 6 BIOBASE GmbH, Wolfenbuettel, Germany Cyclonet — AN INTEGRATED DATABASE ON CELL CYCLE REGULATION AND CARCINOGENESIS 34 Miginsky D.S.1,3*, Sokolov S.A.2, Labuzhsky V.V.2, Nikitin A.G.2,3 1 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia 2 A.P. Ershov Institute of Informatics Systems SB RAS 3 Novosibirsk State University, Novosibirsk, Russia OBJECT-ORIENTED APPROACH TO BIOINFORMATICS SOFTWARE RESOURCES INTEGRATION Sarsenbaev K.N. Institute of Radiation Safety and Ecology, Kurchatov, Kazakhstan USE MOLECULAR MARKERS FOR DIFFERENTIATION POPULATIONS OF STIPA CAPILLATA GROWING IN THE REGIONS WITH HIGH CHRONICAL DOSES OF -RADIATION Suslov V.V1*, Sergeev M.G.2, Yurlova N.I.3, Miginsky D.S.1,2 1 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia 2 Novosibirsk State University, Novosibirsk, Russia 3 Institute of Animal Systematics and Ecology, SB RAS, Novosibirsk, Russia THE ONTOLOGY OF ECOSYSTEMS 35 COMPUTER DEMONSTRATIONS (BY SECTIONS) Computer demos will be available during the entire conference day for the appropriate sections Munday, July 17 COMPUTATIONAL STRUCTURAL AND FUNCTIONAL GENOMICS AND TRANSCRIPTOMICS Vishnevsky O.V.1,2*, Konstantinov Yu.M.3 1 Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia 2 Novosibirsk State University, Novosibirsk, Russia 3 Siberian Institute of Plant Physiology and Biochemistry SB RAS, Irkutsk, Russia ANALYSIS OF THE NUCLEOTIDE CONTEXT OF HIGHER PLANT MITOCHONDRIAL mRNA EDITING SITES Thursday, July 20 COMPARATIVE AND EVOLUTIONARY GENOMICS AND PROTEOMICS Chupov V.S.*, Machs E.M. Komarov Botanical institute RAS, St.Petersburg, Russia VARIATIONS IN NUCLEOTIDE COMPOSITION OF THE REGION ITS1-5.8S RDNA-ITS2 IN EVOLUTIONARY ADVANCED AND EVOLUTIONARY STATIC BRANCH OF THE MONOCOTYLEDONOUS PLANTS NEW APPROACHES TO ANALYSIS/MODELING OF BIOMOLECULAR DATA AND PROCESSES Kolpakov F.1,2,*, Poroikov V.3, Sharipov R.4,2,1, Milanesi L.5, Kel A.6 1 Design Technological Institute of Digital Techniques, SB RAS, Novosibirsk, Russia 2 Institute of Systems Biology, Novosibirsk OOO, Russia 3 Institute of Biomedical Chemistry, RAMS, Moscow, Russia 4 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia 5 Institute of Biomedical Technologies, Segrate (MI), Italy 6 BIOBASE GmbH, Wolfenbuettel, Germany Cyclonet — AN INTEGRATED DATABASE ON CELL CYCLE REGULATION AND CARCINOGENESIS Kolpakov F.1,2*, Puzanov M.1,2, Koshukov A.1,2 1 Institute of Systems Biology OOO, Novosibirsk, Russia 2 Design Technological Institute of Digital Techniques SB RAS, Novosibirsk, Russia BioUML: VISUAL MODELING, AUTOMATED CODE GENERATION AND SIMULATION OF BIOLOGICAL SYSTEMS 36 Friday, July 21 Saturday, July 22 COMPUTATIONAL SYSTEMS BIOLOGY Fadeev S.I.1*, Korolev V.K.1, Gainova I.A.2, Medvedev3A.E. 1 Sobolev Institute of Mathematics SB RAS, Novosibirsk, Russia 2 Khristianovich Institute of Theoretical and Applied Mechanics SB RAS, Novosibirsk, Russia 3 Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia THE PACKAGE STEP+ FOR NUMERICAL STUDY OF AUTONOMOUS SYSTEMS ARISING WHEN MODELING DYNAMICS OF GENETIC-MOLECULAR SYSTEMS Kolpakov F.1,2*, Sharipov R.3,1,2, Cheremushkina E.2, Kalashnikova E.3 1 Institute of Systems Biology OOO, Novosibirsk, Russia 2 Design Technological Institute of Digital Techniques, SB RAS, Novosibirsk, Russia 3 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia BioPATH – A NEW APPROACH TO FORMALIZED DESCRIPTION AND SIMULATION OF BIOLOGICAL SYSTEMS Rudenko V.M.*, Korotkov E.V. Center of Bioengineering RAS, Moscow, Russia CONSTRUCTION OF THE HCV-HEPATOCYTE SYSTEM MODEL Sharipov R.N.1,2,3*, Kolpakova A.F.4 1 Institute of Cytology and Genetics, SB RAS, Novosibirsk, Russia 2 Design Technological Institute of Digital Techniques SB RAS, Novosibirsk, Russia 3 Institute of Systems Biology OOO, Novosibirsk, Russia 4 State Research Institute for Medical Problems of the North, SB RAMS, Krasnoyarsk, Russia FORMALIZED DESCRIPTION OF NF-B PATHWAY, ITS ROLE IN INFLAMMATION, INHIBITION OF APOPTOSIS, CARCINOGENESIS, AND WAYS OF INACTIVATION FOR PREDICTION OF NEW TARGETS FOR ANTI-INFLAMMATORY AND ANTI-CANCER TREATMENT 37