Modeling the Cell Cycle
advertisement

Created by: Claudia Neuhauser
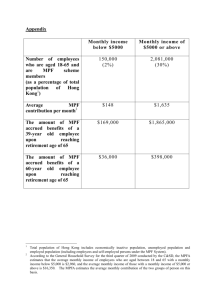
Worksheet 11: Cell Cycle
Modeling the Cell Cycle
Norel and Agur Model (1991) (Science 251: 1076-1078)
This is the simplest model of the interaction between the maturation promotion factor (MPF) and
cyclin. It is a phenomenological model that describes the alternation between interphase and
mitosis. The assumptions are (1) cyclin synthesis activates MPF, (2) MPF activity is
autocatalytic, (3) MPF is inactivated by an inactivase that is constant, (4) cyclin synthesis occurs
at a constant rate, and (5) cyclin is degraded by MPF. A set of equations describing these
assumptions was developed by Norel and Agur.
dM
gM
eC fCM 2
dt
M 1
dC
iM
dt
where M denotes the concentration of MPF and C the concentration of cyclin. The parameter
values are e 3.5, f 1.0, g 10.0, i 1.2 . Here is the Matlab code:
% Solving the Norel and Agur Model na.m
e=3.5;
f=1.0;
g=10.0;
i=1.4;
options=[];
[t,y]=ode23(@nafunc,[0,10],[1e-3 0],options,e,f,g,i);
m=y(:,1);
c=y(:,2);
x=0:0.01:5;
f=g.*x./((x+1).*(e+f.*x.^2));
y=0:0.01:1.4;
z=0.*y+i;
subplot(1,2,1), plot(x,f,'k',z,y,'b');xlabel('MPF
concentration');ylabel('cyclin concentration');title('ZNGI');
legend('dM/dt=0','dC/dt=0');
subplot(1,2,2), plot(t,m,'k',t,c,'b');
xlabel('time');ylabel('concentration');
legend('[M]','[C]');
Worksheet 11: Cell Cycle
% Norel and Agur (1991) Science 251: 1076 Function nafunc.m
function dydt = f(t,y,e,f,g,i)
% m=y(1), c=y(2)
dydt=zeros(2,1);
dydt(1) = e*y(2)+f*y(2)*y(1)^2-g*y(1)/(y(1)+1);
dydt(2) = i-y(1);
The model output shows oscillations of MPF and cyclin, as observed in cell cycles.
3
[M]
[C]
2.5
concentration
2
1.5
1
0.5
0
0
1
2
3
4
5
time
6
7
8
9
10
Figure 1: Dynamics of the Norel and Agur model.
The behavior of this model depends on the rate at which cyclin is produced (constant rate i). As
the rate i increases from small to large values, the behavior changes from exhibiting a point
equilibrium to sustained oscillations.
2
Worksheet 11: Cell Cycle
ZNGI
1.4
1.4
[M]
[C]
1.2
1.2
1
1
0.8
0.8
concentration
cyclin concentration
dM/dt=0
dC/dt=0
0.6
0.6
0.4
0.4
0.2
0.2
0
0
1
2
3
MPF concentration
4
0
5
0
2
4
6
8
10
time
Figure 2: Behavior when i=0.7.
As we increase i, we get oscillations.
ZNGI
1.4
7
dM/dt=0
dC/dt=0
1.2
5
concentration
cyclin concentration
1
0.8
0.6
0.4
4
3
2
1
0.2
0
[M]
[C]
6
0
0
2
4
MPF concentration
6
-1
0
5
time
Figure 3: Behavior when i=1.4.
3
10
Worksheet 11: Cell Cycle
Graphical Approach to Equilibria and Stability
Systems of two differential equations can be analyzed graphically. Let’s consider again the
system
dx
f ( x, y )
dt
dy
g ( x, y )
dt
The first step is to find the equations of the zero isoclines, which are defined as the set of points
that satisfy
0 f ( x, y )
0 g ( x, y )
Each equation results in a curve in the x-y space. Equilibria occur where the two isoclines
intersect (Figure 4).
y
f ( x, y ) 0
Equilibrium
g ( x, y ) 0
x
Figure 4: Zero isoclines corresponding to the two differential equations. Equilibria occur where the isoclines
intersect.
It is sometimes possible to determine stability of an equilibrium graphically based on the sign
structure of the corresponding Jacobian matrix. To determine the sign structure of the
corresponding Jacobian matrix, we redraw Figure 1 and include the signs of the two functions f
and g as well (Figure 5).
We can now determine the sign structure of the Jacobian matrix by following the two
arrows through the equilibrium and recording how f (respectively, g) changes as either x or y
changes. This will determine the signs of the partial derivatives in the Jacobian matrix.
4
Worksheet 11: Cell Cycle
f
first. When we follow the horizontal arrow through the equilibrium
x
point, we move from a region where f is positive to a region where f is negative, implying that f
f
0 . The other entries of the Jacobian matrix can be
is decreasing as x increases. Thus,
x
found similarly, which yields
Let’s look at
J ( xˆ, yˆ )
y
f 0
g0
f 0
g0
g ( x, y ) 0
f 0
g 0
f 0
g 0
g ( x, y ) 0
x .
Figure 5: The two isoclines divide the plane into four regions. Each region is labeled according to the signs of the
two functions f and g.
We need the following result from linear algebra:
Given a 2 2 matrix A. Both eigenvalues of A have negative real parts if
det( A) 0 and trace ( A) 0
For the matrix J ( xˆ, yˆ )
, we find det( A) 0 and trace ( A) 0 . Thus both eigenvalues
have negative real parts.
5
Worksheet 11: Cell Cycle
This method does not always work as it might not be possible to determine the signs of
the determinant or the trace based on the limited information provided by the sign structure of
the Jacobian matrix.
Back to the Norel and Agur Model
The vertical zero net growth isocline (ZNGI) comes from C 0 . The other ZNGI comes from
M 0 . To the left of the C 0 ZNGI, C 0 , to the right, C 0 . Below the M 0 ZNGI,
M 0 , above M 0 . This allows us to find the signs of the entries in the Jacobi matrix at the
point equilibrium.
Looking at the left panel in Figure 2, we can derive the signs in the Jacobi matrix at the
point equilibrium, namely
J (M , C )
0
Hence, the trace is negative and the determinant is positive, implying that both eigenvalues have
negative real parts. It follows that if the vertical line intersects the other isocline to the left of the
maximum, then the point equilibrium is stable. This is consistent with the behavior exhibited in
the right panel of Figure 2.
Looking at the left panel in Figure 3, we find for the signs in the Jacobi matrix evaluated
at equilibrium
J (M , C )
0
It follows that the trace and the determinant are positive. Therefore, the point equilibrium is
unstable. This graphical analysis allows us to determine the stability of the point equilibrium. To
show the existence of a limit cycle (sustained oscillations) is more difficult and beyond what we
can do here. This is consistent with the behavior exhibited in the right panel of Figure 3.
Goldbeter Model (1991) (PNAS 88: 9107-9111)
The Goldbeter model for the mitotic oscillator describes the dynamics of cyclin concentration
(C), the fraction of active cdc2 kinase (M) and the fraction of active cyclin protease (X). Cyclin is
produced at a constant rate and decays at a constant rate and a rate that increases with the
fraction of active cyclin protease. Cyclin in turn triggers the transformation of inactive into
active cdc2 kinase, which activates cyclin protease. This generates a negative feedback loop that
results in oscillations of cyclin and cdc2 kinase. The dynamics are described by the following set
of differential equations.
6
Worksheet 11: Cell Cycle
dC
C
1 d X
kd C
dt
Kd C
dM
(1 M )
M
V1
V2
dt
K1 (1 M )
K2 M
dX
(1 X )
X
V3
V4
dt
K 3 (1 X )
K4 X
C
VM 1 and V3 MVM 3 . The parameters in the model are d 0.25 , i 0.025 ,
KC C
K d 0.02 , kd 0.01 , VM 1 3 , V2 1.5 , VM 3 1 , V4 0.5 , KC 0.5 , and Ki 0.005 or10
( i 1 4 ).
where V1
Task 1
Code up the Goldbeter model in Matlab and generate the figure below with the parameters given
above.
concentration
0.8
[C]
[M]
[X]
0.6
0.4
0.2
0
0
10
20
30
40
50
time
M
Xeq
0.5
0
70
80
90
100
1
0.5
0
0
0.5
1
1.5
Ceq
2
2.5
3
0.8
0.8
0.6
0.6
0.4
0.4
X
Meq
1
60
0.2
0
0.1
0
0.5
1
1.5
Meq3
2
2.5
3
0.2
0.2
0.3
0.4
0.5
0
0.6
C
7
0
0.2
0.4
M
0.6
0.8
Worksheet 11: Cell Cycle
Task 2
How were the two panels in Figure 2 in the Goldbeter paper generated?
Tyson Model (1991) (PNAS 88: 7328-7332)
The interactions between cyclin and cdc2 are modeled in more detail in the Tyson model. Cyclin
is synthesized de novo and combines with phosphorylated cdc2 to form an inactive maturation
promotion factor (MPF) complex upon phosphorylation. This complex becomes activated
through autocatalytic dephosphorylation (the active MPF complex acts as a catalyst). Active
MPF triggers cell division if MPF is present in sufficient quantities. Active MPF breaks down
into its two components, cyclin and cdc2. Cyclin degrades and cdc2 is phosphorylated. These
processes are described by the following set of ordinary differential equations.
d [C 2]
k6 [ M ] k8 [~ P][C 2] k9 [CP]
dt
d [CP ]
k3 [CP][Y ] k8 [~ P][C 2] k9 [CP]
dt
d [ pM ]
k3 [CP][Y ] [ pM ]F ([ M ]) k5[~ P][ M ]
dt
d[M ]
[ pM ]F ([ M ]) k5 [~ P][ M ] k6 [ M ]
dt
d [Y ]
k1[aa ] k2 [Y ] k3[CP ][Y ]
dt
d [YP ]
k6 [ M ] k7 [YP ]
dt
Task 3
Relate the system of differential equations to Figure 1 in Tyson (1991).
Task 4
There is a conserved quantity in this model. Find it and explain its biological meaning.
Homework (due Thursday, December 6)
Read Tyson, Chen and Novak 2001. Network Dynamics and Cell Physiology. Nature
Reviews.
Homework (due Tuesday, December 11)
Complete Tasks 1-4
8