PDF - Thinking Writing
advertisement

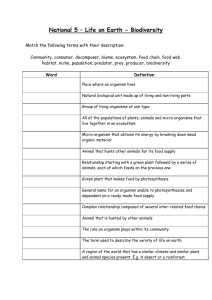
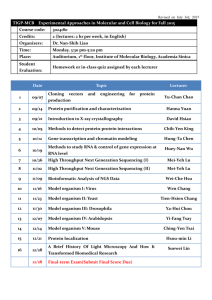
Unknown Lab Report III Identification of Indigenous Unknowns Introduction: In a scientific field with so much genetic variation and rapidly evolving species, it has become increasingly difficult to correctly identify unknown microorganisms. With this increase in difficulty, we have also seen an increase in the importance of a correct identification. Because of the many different treatments needed to cure bacterial infections and diseases, it has become crucial that the organism affecting the patient be correctly identified so that the proper treatment can be prescribed. The experiment conducted was used to demonstrate this concept, and how a clinical microbiologist might go about properly isolating and identifying an unknown organism from various body sites. In addition to this objective, the experiment was also carried out to show just how colonized the human body is with bacteria, comprised of both normal microbiota and opportunistically pathogenic bacteria. Methods: In order to isolate individual bacterial species from various body sites, many steps have to be taken. Samples were recovered from the oropharynx (back of the throat), the nares (inside of the nose), and the back surface of the hand. To recover the bacteria from the back of the throat, a sterile swab was used to collect the sample and then was used to heavily inoculate the first area of a blood agar plate. After using the swab, an inoculating loop was used to finish streaking the blood agar for isolation (1). Bacteria were recovered from the inside of the nose using a damp sterile swab which was then used to heavily inoculate the first area of a new blood agar plate. After the swab was used, an inoculating loop was again used to streak the rest of the blood agar plate for isolation (1). Finally, to recover bacteria from the hand, a wet sterile swab was used. Unlike how the previous plates were struck for isolation, the swab with recovered bacteria was used to spread the material over an entire blood agar plate, where the plate was rotated 120ºand swabbed from top to bottom. The plate was rotated 120º like this twice more, again swabbing from top to bottom after each turn. These three plates were then incubated at 3537ºC for 48 hours (1). After incubation, the three plates were analyzed, and a total of 5 different colonies of noticeably different bacteria were selected from the plates, with at least one colony coming from each body location’s plate. Each of these colonies was sub-cultured onto another blood agar plate and a tryptic soy agar (TSA) plate and each were struck for isolation. These plates were then incubated at 35-37ºC for 28 hours. After incubation, the plates were analyzed to confirm isolation. Before individual tests were done on each of the three unknown organisms selected, macro and microscopic observations along with a few tests and plating on more selective media were completed for all 5 organisms to give a better idea as to their nature. The microscopic observations were taken from a Gram stained slide of each organism. In a Gram stain, two different stains are used to allow for organization of organisms based on their cell wall makeup. Crystal violet, a purple dye, was first placed on the slide with the organism already inoculated and was left to sit for one minute. After the slide was rinsed with water, Gram’s iodine was added as a mordant and was left to sit for one minute. After the slide was again rinsed with water, acetone was quickly washed over the slide for no more than 10 seconds. After rinsing the slide once again with water, the safranin counterstain was placed on the slide and was left to sit 1 for 30 seconds. Once stained, the slide was dried with paper towels and was examined under high power (1000X) using a bright field microscope (2). A catalase test was the first performed. A drop of hydrogen peroxide was placed on the organism which had been placed on a glass slide, and the production of oxygen or lack thereof was recorded (3). To confirm the Gram stain results, each of the organisms was also grown on the selective media colistin, nalidixic acid (CNA) plate and MacConkey agar plate. CNA plates are selective for the growth of Gram positive species of bacteria, while MacConkey agar is selective for the growth of Gram negative species of bacteria and differential in that the agar indicates lactose fermentation or lack thereof (4). These media need not be struck for isolation, so the plates were inoculated in a loose zigzag pattern. After these tests were completed, 3 of the 5 unknown organisms were chosen to be identified. From this point on, these 3 organisms were continually sub-cultured on TSA every 23 weeks if the organism was able to grow on the agar or blood agar every 1-2 weeks if TSA growth was not possible (as was the case for one of the organisms chosen). In order to create a plan of action for individual testing, various dichotomous keys from a textbook (Chess, 2009) were used. Because some common staphylococcal species are anaerobic and others are strictly aerobic, organisms that were hypothesized to be staphylococcal had to be distinguished in this regard through the use of anaerobic Gas-Pak chambers. Once the aerobic or anaerobic nature of the staphylococcal species was established, the dichotomous keys provided were followed to correctly identify the organism. At different times throughout the individual organism testing, phenyl red sugar broths were used. Glucose, sucrose, maltose, and mannitol phenyl red sugar broths were all used in the process of identification. For these tests, a loopful of colony was placed in the broth and was incubated for approximately 48 hours (4). An arginine decarboxylase test was also used in various places during individual organism testing. Two tubes of Moeller decarboxylase medium were inoculated, one containing the arginine and the other one used as a control containing no amino acids. The control tube is necessary because the enzymes in the broth must be in a low enough pH to become active, so the broth includes pH indicator to determine if the organism is viable and that the pH of the medium containing the amino acids has become more acidic (indicating that the enzymes are activated). A 0.5 inch layer of mineral oil was added to the top of both tubes after inoculation and before incubation (4). Another test used was the nitrate test. A tube of broth was inoculated with a colony and was incubated for 48 hours. After incubation, 1 mL each of nitrate test reagents A and B were added to the broth. If no color change has occurred, a very small amount of zinc is added to confirm a negative test or indicate a positive result not tested for in the original nitrate broth (4). Susceptibility to penicillin was also tested. A blood agar plate was inoculated using a swab to completely cover the plate, and a filter disk containing penicillin was placed on the center of the agar. The plate was incubated for 48 hours (4). Results: 2 Organism #1 (as referred to in Table 1) was from the skin swab and a stock was maintained throughout the experimentation on blood agar. This organism was a Gram positive cocci with a cell size of approximately 1 µm, as indicated by a purple-stained cell appearance. It displayed a short chain and/or clustered arrangement under magnification (see Gram stain sketch below). Macroscopically, the organism grew in very small, punctiform colonies with an entire edge and a slight cream coloration. Organism #1 was α- hemolytic on the blood agar, meaning that it is partially hemolytic, as indicated by a greenish color in the blood agar surrounding the colonies. In the catalase test, this organism tested negative, as indicated by no oxygen bubbles forming on the colony placed on the glass slide after a drop of hydrogen peroxide was placed on the colony. This organism did not grow on the MacConkey agar and grew very poorly on the CNA plate. Susceptibility to penicillin was tested, and resulted in no zone of inhibition of growth, indicating resistance to penicillin. A mannitol phenyl red broth was also used on organism #1, and a negative result was seen as indicated by no color change from red to yellow. The final test used was an arginine decarboxylase test. The control tube turned yellow, indicating that the organism was viable and that the enzymes were activated. The tube with the arginine present gave a negative result, as indicated by a yellow color change in the broth. Organism #2 (as referred to in Table 1) was from the nares swab and a stock was maintained on a TSA plate for the remainder of the experimental period. This organism was a Gram positive cocci with a cell size of approximately 1 µm. It displayed clustered arrangement under magnification (see Gram stain sketch below). Macroscopically, organism #2 had 2-3 mm, round, entire edged colonies with a white or cream colored appearance. This organism was unable to grow on the MacConkey but displayed full growth on the CNA plate, confirming its positive Gram stain result. The catalase test yielded a positive result, as indicated by oxygen bubbles forming after a drop of hydrogen peroxide was placed on the colony. On the blood agar, organism #2 displayed γ-hemolysis, meaning that it was not hemolytic, as indicated by no change in color or density of red color in the agar. As a white-colonied, γ-hemolytic, catalasepositive organism, organism #2 was determined to likely be staphylococcal. In order to follow the dichotomous keys provided, this staphylococcal species had to be tested for anaerobic growth. The organism was able to grow in both the anaerobic chamber and aerobically, making this organism fall under the anaerobic staphylococcal species dichotomous key. Sucrose and maltose phenyl red broths were all used to help identify this organism. Both broths tested positive, meaning that the organism was able to ferment these sugars, as indicated by a color change from red to yellow in the broths. A nitrate test was also carried out. After the reagents were added, no color change occurred, indicating that nitrites were not present. With this negative result, zinc was added. A red color change occurred after the addition of zinc, indicating that zinc had reduced the nitrates to nitrites, giving a true negative result. Finally, an arginine decarboxylase broth was used, and yielded a yellow control tube and a yellow arginine tube result, giving a negative result. Organism #3 (as referred to in Table 1) was also from the skin culture, and was maintained on TSA plates for testing throughout the experimental period. This organism was a Gram positive cocci with a cell size of approximately 1-1.5 µm, and was arranged in characteristic tetrads and clusters (see Gram stain sketch below). Macroscopically, organism #3 had a small, circular, entire edged colonies with bright yellow pigmentation. On the blood agar, the organism displayed β-hemolysis, meaning that the organism is fully hemolytic, as indicated by a clearing of red color around the colonies, where the plate has become considerably translucent around the colonies. Organism #3 was unable to grow on the MacConkey agar plate 3 and displayed full growth on the CNA plate, confirming its positive gram stain result. The organism was very catalase positive, and produced a large amount of oxygen bubbles. Based on its catalase-positive, gram-positive cocci, tetrad cell arrangement, and bright yellow colony characteristics, the organism was determined to be a Micrococcus species, which allowed for further dichotomous key use. A glucose phenyl red broth was used and the organism tested negative for glucose fermentation, as indicated by no color change in the broth. Gram stain results: Organism #1 (1000X magnification) Organism #2 (1000X magnification) Organism #3 (1000X magnification) Table 1. Results of various macro and microscopic observations and tests performed Test/Observation Organism #1 Organism #2 Organism #3 Colony color clear/white cream/white bright yellow Colony size punctiform medium 2-3 mm small 1-mm Colony shape/margin round/entire round/entire round/entire Growth maintained blood agar TSA TSA on: Gram stain + + + Cell shape cocci cocci cocci Cell size 1 µm 1 µm 1-1.5 µm Cell arrangement small chains/clusters clusters tetrads/clusters Blood agar hemolysis α γ β MacConkey agar growth CNA growth weak + + + Catalase test + ++ P (penicillin) disk resistant (not tested) (not tested) Phenyl red sucrose (not tested) + (not tested) Phenyl red mannitol (not tested) (not tested) Phenyl red maltose (not tested) + (not tested) Phenyl red glucose (not tested) (not tested) Nitrate reduction (not tested) (not tested) Arginine (not tested) decarboxylase Discussion: 4 With the amount of different species of organisms that normally inhabit the body, it becomes almost essential to use dichotomous keys in order to create a logical approach to the proper identification of an organism. A few standard tests were completed on all three of the organisms to give some direction and to determine which dichotomous key (Chess, 2009) needed to be used for each unknown. Organism #1 was a gram positive, catalase-negative cocci, which is characteristic of streptococcal species, as confirmed by the first dichotomous key. Two dichotomous keys remained for streptococcal species, one for pyogenic (pus forming) streptococcal species and one for oral streptococcal species. Because this organism was an unknown, neither the oral or pyogenic nature of the organism could be determined, so tests were first carried out following both keys. The α-hemolysis of organism #1 narrowed the possible organism identity down to just four organisms, two from each dichotomous key. The negative result seen in the phenyl red mannitol broth allowed for one of the pyogenic, α-hemolytic organisms to be ruled out as a possible identification. A negative result means that organism #1 is unable to ferment mannitol (4). This left just Streptococcus pneumonia (S. pneumonia) under the pyogenic Streptococcus dichotomous key and the two organisms from the oral, α-hemolytic Streptococcus dichotomous key. Because S. pneumonia is susceptible to penicillin, a result of no inhibition of organism #1 in the presence of a penicillin disk ruled out S. pneumonia, leaving just Streptococcus mitis (S. mitis) and Streptococcus sanguis (S. sanguis) as the possible identities of the unknown. Both organisms are mannitol negative, confirming that these organisms are still viable identities of organism #2 (4). A negative result in the arginine decarboxylase test revealed that organism #1’s identity is S. mitis. Please refer to the dichotomous keys (Chess, 2009) attached for better understanding of the steps taken to reach this conclusion. Streptococcus mitis is an opportunistic pathogen that is normal flora of the oral cavity where it colonizes hard dental tissue and mucous membranes. This organism commonly causes bacterial endocarditis, or inflammation of the inner layer of the heart. S. mitis does not form spores and is a mesophile, growing best at temperatures between 30ºC and 35ºC (5). Because of its ubiquity in the oral cavity of humans, it is very possible to find this organism on the skin of the hand, where this organism was originally recovered from. Organism #2 was determined to be staphylococcal in nature based on a few test results and observations. Its positive catalase test result indicates that this organism is able to breakdown hydrogen peroxide into water and oxygen. Based on its positive Gram stain, its catalase positive nature, its coccus cell shape, and irregular cell arrangement, this organism could be narrowed down to being either a staphylococcal or planococcal species. The two genera are distinguished by motility, where Staphylococcus species are nonmotile, but because of the overwhelmingly larger amount of staphylococcal species that come into contact with the human body than planococcal species, it could be assumed that the species is staphylococcal. Once this was established, a test for anaerobic growth was used to determine which staphylococcal dichotomous key should be used. This organism was able to grow in the anaerobic chamber and in the aerobic environment, meaning that this organism does not require oxygen for growth but can use oxygen when available. This type of organism is known as a facultative anaerobe (4). Using the anaerobic staphylococcus dichotomous key for sucrose fermentation-positive organisms, the possible identity of the organism was narrowed down to 10 organisms. The positive phenyl red sucrose test indicates that this organism is able to ferment sucrose. A maltose test was used next, and a positive result, meaning that organism #2 can ferment maltose, allowed for the list of possibly identities of the organism to be narrowed down to 6. A negative nitrate result was seen for this organism, which was confirmed through the production of a red color 5 once zinc was added to the nitrate broth containing the organism and the 2 nitrate reagents. This result means that zinc (and not the organism) reduced the nitrates still remaining in the broth, indicating that organism #2 cannot reduce nitrates at all (4). This negative result left only two possible organism identities, Staphylococcus warneri (S. warneri) and Staphylococcus saprophyticus (S. saprophyticus). An arginine decarboxylase test distinguishes the two from one another, and a negative arginine decarboxylase result allowed for the conclusive identification of organism #2 as S. saprophyticus. Please refer to the attached dichotomous keys (Chess, 2009) for better understanding of the steps taken to come to this conclusion. S. saprophyticus is a facultative anaerobe that commonly causes cystitis, or urinary bladder inflammation, and/or uncomplicated urinary tract infections in both males and females, causing approximately 10-20% of all urinary tract infections in sexually active women. S. saprophyticus is not normally present on the body, so the recovery of this organism from the nares creates a source of debate (6). The most likely reason behind this recovery is that the organism somehow became trapped in the nasal mucosa, and was recovered quickly after being trapped since this organism does not normally grow in or on the body. Organism #3 was determined to be part of the Micrococcus genus based on a few observations and test results. This gram positive, catalase positive organism was narrowed down to being micrococcal, planococcal, or staphylococcal in nature based on these results alone. A tetrad arrangement of the cells is very characteristic of Micrococcus species, and the bright yellow colony color is characteristic of some very common Micrococcus species, so the identity of the organism was able to be narrowed down to being one of 9 Micrococcus species. The yellow colony color allowed for the number of possible organisms to be narrowed down to just two bacteria, Micrococcus varians (M. varians) and Micrococcus luteus (M. luteus). A phenyl red glucose test differentiates the two, where M. luteus is unable to ferment glucose. The negative result of the phenyl red glucose broth makes it possible to conclusively identify organism #3 as Micrococcus luteus. Please refer to the attached dichotomous keys (Chess, 2009) for a better understanding of the steps taken to come to this conclusion. M. luteus is an obligate aerobe part of the normal flora of human skin, and can also colonize the mouth, mucosae, oropharynx, and upper respiratory tract. It is an opportunistic pathogen that can cause septic shock, pneumonia, and urinary tract infections in immunecompromised individuals (7). Because this organism is part of the normal skin flora, the fact that this organism was recovered from the skin of the hand is not surprising. Works Cited 6 1. Eggers, C. Lab II: Isolation of Indigenous Microorganisms Laboratory Handout. n.d. 2. MicrobiologyBytes. Gram Stain. 13 April 2011 <http://www.microbiologybytes.com/video/Gram.html>. 3. Georgia Highlands College. Catalase Test. 3 February 2010. 3 March 2011 <http://www.highlands.edu/subwebs/labcoordinator/Microbiology/catalase_coagulase_test.htm>. 4. Winn, Jr., Washington, et al. Koneman's Color Atlas and Textbook of Diagnostic Microbiology. Baltimore: Lippincott Williams and Wilkins, 2006. 5. MicrobeWiki. Streptococcus mitis. 29 April 2011. 5 May 2011 <http://microbewiki.kenyon.edu/index.php/Streptococcus_mitis>. 6. MicrobeWiki. Staphylococcus saprophyticus. 10 August 2010. 5 May 2011 <http://microbewiki.kenyon.edu/index.php/Staphylococcus_saprophyticus>. 7. The Lab Rat. Micrococcus luteus. 2005. 5 May 2011 <http://www.thelabrat.com/restriction/sources/Micrococcusluteus.shtml>. 8. Chess, Barry. Dichotomous Keys taken from Laboratory Applications in Microbiology. 2009. 7