Why do models matter? - molecularevolution.org
advertisement

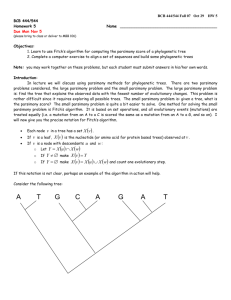
Why do models matter? • Model-based methods including ML and Bayesian inference (typically) make a consistent estimate of the phylogeny (estimate converges to true tree as number of sites increases toward infinity) ... even when you’re in the “Felsenstein Zone” A C (Felsenstein, 1978) B D In the Felsenstein Zone Proportion Correct 1 0.8 parsimony 0.6 ML-GTR 0.4 0.2 0 0 5000 Sequence Length Sequence Length Simulation model = GTR 10000 Why do models matter (continued)? • Parsimony is inconsistent in the Felsenstein zone (and other scenarios) • Likelihood is consistent in any “zone” (when certain requirements are met) But this guarantee requires that the model be specified correctly! Likelihood can also be inconsistent if the model is oversimplified • Real data always evolve according to processes more complex than any computationally feasible model would permit, so we have to choose “good” rather than “correct” models What is a “good” model? • A model that appropriately balances fit of the data with simplicity (parsimony, in a different sense) i.e., if a simpler model fits the data almost as well as a more complex model, prefer the simpler one 100 120 B 80 80 60 B B y 40 y B 40 B 25 B B -40 B B 0 B B B B 0 B B 20 0 B 50 x 75 100 -80 0 25 50 x 75 100 y = - 330 +134x - 15.5x 2 +0.816x 3 y =1.30 + 0.965x - 0.0225x 4 + 0.000335x 5 (r 2 = 0.963) - 0.00000255x 6 +0.00000000777x 7 (r 2 =1.000) “The Principle of Parsimony” in the world of statistics • Burnham and Anderson (1998): Model Selection and Inference – Parsimony lies between the evils of underfitting and overfitting. The concept of parsimony has a long history in in the sciences. Often this has been expressed as “Occam’s razor”—shave away all that is not necessary. Parsimony in statistics represents a tradeoff between bias and variance as a function of the dimension of the model. A good model is a balance between under- and over-fitting. Why models don’t have to be perfect Assertion: In most situations, phylogenetic inference is relatively robust to model misspecification, as long as critical factors influencing sequence evolution are accommodated Caveat: There are some kinds of model misspecification that are very difficult to overcome (e.g., “heterotachy”) E.g.: A C B D Half of sites A C D B Other half Likelihood can be consistent in Felsenstein zone, but will be inconsistent if a single set of branch lengths are assumed when there are actually two sets of branch lengths (Chang 1996) GTR Family of Reversible DNA Substitution Models (general time-reversible) GTR 3 substitution types (transversions, 2 transition classes) Equal base frequencies TrN SYM (Tamura-Nei) 3 substitution types (transitions, 2 transversion classes) 2 substitution types (transitions vs. transversions) (Hasegawa-Kishino-Yano) (Felsenstein) HKY85 F84 Equal base frequencies Single substitution type (Felsenstein) K3ST 2 substitution types (transitions vs. transversions) K2P F81 Equal base frequencies (Kimura 3-subst. type) (Kimura 2-parameter) Single substitution type JC Jukes-Cantor Among site rate heterogeneity equal rates? Lemur Homo Pan Goril Pongo Hylo Maca • …TTACATCATCCA …TTACATCCTCAT …TTACATCCTCAT …CCCACGGACTTA …GCAACCACCCTC …TGCAACCGTCCT …CGCAACCATCCT Some sites extremely unlikely to change due to strong functional or structural constraint (Hasegawa et al., 1985) Gamma-distributed rates – • TTGCATCATCCA TTGCATCATCCA TTACGCCATCCA TTACGCCATCCA TTACGCCATCCT TTACATTATCCG TTACATTATCCG Proportion of invariable sites – • AAGCTTCATAG AAGCTTCACCG AAGCTTCACCG AAGCTTCACCG AAGCTTCACCG AAGCTTTACAG AAGCTTTTCCG Rate variation assumed to follow a gamma distribution with shape parameter α Site-specific rates (another way to model ASRV) – Different relative rates assumed for pre-assigned subsets of sites Modeling ASRV with gamma distribution 0.08 α=200 Frequency 0.06 α=0.5 α=2 0.04 α=50 0.02 0 0 1 2 Rate …can also include a proportion of “invariable” sites (pinv) Performance of ML when its model is violated Tree α = 0.5, pinv=0.5 α = 1.0, pinv=0.5 1 1 1 0.9 0.9 0.9 0.8 0.8 0.7 GTRig 0.6 HKYig 0.4 HKYg GTRi 0.3 HKYi 0.6 0.1 0.1 0.4 0.3 HKYer 0.2 parsimony 0.1 0 0 100 1000 10000 100000 100 1000 10000 100 100000 1 1 1 0.9 0.9 0.9 0.8 0.8 0.7 GTRig GTRig 0.6 HKYig GTRg 0.5 GTRg 0.5 0.2 parsimony 0 100 0.4 HKYg GTRi 0.3 HKYi GTRer 0.2 HKYer 0.1 0.8 0.7 0.5 GTRer HKYer 0.1 10000 100000 100 0.4 GTRg HKYg GTRi 0.3 HKYi GTRer 0.2 parsimony HKYer parsimony 0.1 0 0 1000 1000 10000 100000 100 1 1 1 0.9 0.9 0.9 0.8 0.8 0.7 GTRig HKYig 0.6 GTRg HKYg GTRi HKYi GTRer 0.5 0.4 0.3 0.2 0.1 100 GTRig 0.7 0.6 HKYig 0.6 GTRg 0.3 10000 100000 0.2 0.3 0.2 HKYer 0.1 100 100000 0.4 GTRer parsimony 0 1000 10000 0.5 HKYg GTRi HKYi 0.4 0 1000 0.8 0.7 0.5 HKYer parsimony 100000 HKYig 0.6 0.3 10000 GTRig HKYig 0.4 1000 0.7 0.6 HKYg GTRi HKYi GTRig HYYig GTRg HKYg GTRi HKYi GRTer HKYer parsimony 0.5 GTRer 0.2 0 0.6 GTRi HKYi 0.3 Parsimony 0.7 HKYig HKTg 0.4 HKYer GTRig GTRg 0.5 GTRer 0.2 0.8 0.7 GTRg 0.5 Proportion Correct α = 1.0, pinv=0.2 0.1 0 1000 10000 Sequence Length 100000 100 1000 10000 100000 “MODERATE”–Felsenstein zone α = 1.0, pinv=0.5 1 0.9 0.8 JCer 0.7 JC+G 0.6 JC+I JC+I+G GTRer GTR+G 0.5 0.4 GTR+I 0.3 GTR+I+G parsimony 0.2 0.1 0 100 1000 10000 100000 “MODERATE”–InverseFelsenstein zone 1 0.9 0.8 JCer 0.7 JC+G JC+I JC+I+G 0.6 0.5 GTRer GTR+G 0.4 GTR+I 0.3 GTR+I+G parsimony 0.2 0.1 0 100 1000 10000 100000 Model selection criteria • Likelihood ratio tests δ = −2(ln L0 − ln L1 ) € If model L0 is nested within model L1, δ is distributed as χ2 with degrees-of-freedom equal to difference in number of free parameters • Akaike information criterion (AIC) AICi = −2ln Li + 2K where K is the number of free parameters estimated • Bayesian information criterion (BIC) € BICi = −2ln Li + K ln n where K is the number of free parameters estimated and n is the “sample size” (typically number of sites) €