Cladistics Teacher's Guide: Cladisticules Exercise
advertisement

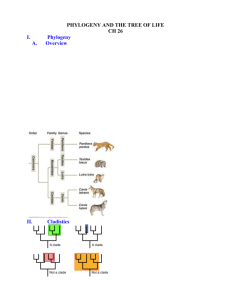
Cladisticules-Teacher’s Guide I. Objectives. The Cladisticules exercise is designed to introduce students (advanced high school or beginning university-level) to the following concepts in phylogeny/evolution: Characters Character states Parsimony Homology Homoplasy Cladograms Ingroup and outgroup Rooting Evolutionary polarity Synapomorphy Symplesiomorphy Monophyly In addition, this exercise allows students to conduct their own simple cladistic analysis on a set of imaginary organisms (Cladisticules). II. Suggestions for teaching the Cladisticules exercise. For optimal impact, we suggest first introducing the concept of shared ancestry and the way that the distribution of homologous characters (homologies) in different organisms can allow us to infer shared ancestry at different levels (more or less inclusive groups). An example that is often useful for this purpose is to discuss the unique characters that unite the group Mammals (e.g. hair, milk)…they are useful for grouping all mammals, but if you wanted to uncover sub-groups within the mammals, you’d need to look at other unique characters that distinguish the various groups. The character hair, for instance, would not be useful for distinguishing the primates from the bats, but the character of flight would be. We have found that one effective technique for presenting the material is to distribute (or present) the Cladisticules exercise in increments. First, provide page 1 (the figure with all the cladisticules ingroup and outgroup), and have the students identify potential homologies (characters) that vary among the creatures shown (the variants being character states within a character). Page 2 identifies some of these characters, so it should not be shown until students have had a chance to work through the exercise of character selection on their own. Once these characters have been identified, have the students fill out the data matrix on page 3 (the completed matrix on page 4 is provided solely for teacher reference). After the students have filled out the data matrix, we suggest introducing the principle of parsimony, that is, the simplest or shortest explanation for the character data is most likely the correct one. This principle is widely used among biologists as a model of how evolution proceeds. In phylogenetic analyses we generally utilize traits that are evolving slowly relative to the age of the group in question so it is reasonable to assume that changes are minimal. In phylogenetic studies, parsimony relates to the number of times a character changes from one state to another for all the taxa on an entire tree. The tree arrangement that minimizes the total number of character state changes is the most parsimonious tree. Thus parsimony does not rely on an assumption that evolution must proceed according to the absolutely shortest route, instead it assumes that nonhomologous change in one character should only inferred if necessary to fit the overall pattern displayed by all characters (see below for an illustration using the cladisticules). it is appropriate at this point to introduce the concept of the outgroup versus the ingroup, and the idea of rooting. We feel that it is important to emphasize that the outgroup is not essential to the process of constructing a cladogram: an outgroup is only used to root the tree (it is perfectly acceptable for many purposes to have an unrooted tree as well). But if we have a good idea of one or more appropriate outgroups (that is, organisms that we know are phylogenetically outside of the group that we want to study), then including outgroups in our analysis is a good idea because ii allows us to root our tree -that is, it allows us to hypothesize where the bottom of the tree is and to see the direction or evolutionary polarity of the changes in character states, from primitive to derived, on the cladogram. Shared derived characters are called synapomorphies, and shared primitive characters are called symplesiomorphies. The distinction is important, since only synapomorphies can be used to define monophyletic taxonomic groups (i.e., groups that contain all and only descendants of a common ancestor). For simple data sets, the parsimony principle can be implemented using set theory. Each character state can be viewed as a set of those organisms possessing that state. For simplicity, we'll use the character state considered to be evolutionarily derived (apomorphic) using outgroup comparison as discussed above. By convention, the inferred plesiomorphic state is coded '0' and the apomorphic state is coded '1' (as in the solution to the data matrix, page 4). Building a tree is equivalent to adding together the sets that are proper subsets of each other. The Venn diagram on page 5 (top), again, can either be provided to the students or used only by the teacher. In our experience presenting this exercise, it is usually most effective to not distribute the Venn diagram to the students, but instead work through the first two or so groupings on a chalk board (working from most inclusive to least inclusive characters, drawing a set boundary around the organisms sharing state '1' in each character), and then allow some time for students to complete Venn diagrams on their own, going back over the correct Venn diagram with the entire class after students have had the opportunity to work on the problem individually. Teacher’s note: the numbers on the diagram (along edge of boxes) represent the characters that group the cladisticules (names inside of boxes) at various levels. How to deal with character conflict? Character #7 unites a group (April, Tanya, and Jane) that is in conflict with the rest of the groupings (this is equivalent to an improper subset in set theory), shown on the bottom of page 5. This is an example of a homoplastic character (homoplasy) -- a character that we initially thought to be a homology, that after our analysis is found to not be a homology (in this case, it could be a convergent similarity due to gender, since it unites the females). It is important to point out that while we try to pick characters that we believe to be homologous, we can only identify non-homologous characters (homoplasies) after our analysis is done (in light of all the evidence gained from the other characters). Once the Venn diagram is completed, have the students attempt to construct a cladogram from it (again, we would suggest withholding the final cladogram on page 7), and then work through the problem together. We show the process on page 6. In producing the final tree, keep in mind that branches can be freely rotated at a node, so the trees on pages 6 and 7 are identical. Be sure to point out to students the power of this sort of evolutionary inference: we can not only reconstruct the branching order (relative recency of common ancestry) but also infer the order of character change. Note that this set-theory approach to building trees would not work for large, complicated data sets, and indeed is not the exact approach that current computer algorithms work to build trees. However, it does illustrate the conceptual framework behind phylogenetic reconstruction accurately, and is the easiest approach for beginners to grasp intuitively. Students should be encouraged to look into the subject further, since the whole area of computerassisted phylogenetic analysis is rapidly developing and being applied to many exciting topics in biology including biogeography, community ecology, behavior, development, public health, and medicine.