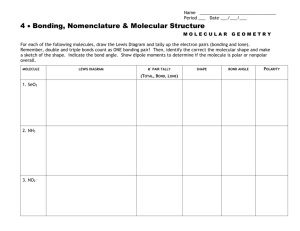
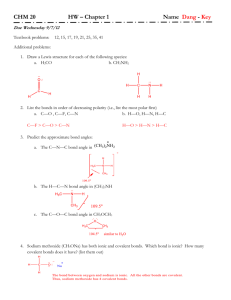
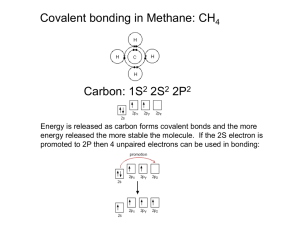
Chapter 9 Molecular Structures Molecular Structures Using
advertisement

Molecular Structures John W. Moore Conrad L. Stanitski Peter C. Jurs C2H6O structural isomers: H H | | .. H–C–C–O .. – H | | H H http://academic.cengage.com/chemistry/moore Chapter 9 Molecular Structures Stephen C. Foster • Mississippi State University m.p./ °C b.p./ °C H H | .. | H–C–O –C–H .. | | H H ethanol dimethyl ether -114 +78 -142 -25 Molecular shape is important! Small structural changes cause large property changes. Using Molecular Models Using Molecular Models Physical models of 3D-structures: Hand-drawn molecules: Going back into the screen ball and stick space filling In the plane of the screen H C H Computer versions: H H Predicting Molecular Shapes: VSEPR Coming out of the screen Predicting Molecular Shapes: VSEPR The Valence Shell Electron Pair Repulsion model is used to predict shapes. Key ideas: 1. e- pairs stay as far apart as possible. • Repulsions are minimized. 2. Molecule shape is governed by the number of Linear Triangular planar Tetrahedral bond pairs and lone pairs present. 3. Treat multiple bonds like single bonds. • Each is a single e- group. 4. Lone pairs occupy more volume than bonds. Triangular bipyramidal Octahedral 1 Predicting Molecular Shapes: VSEPR Shapes that minimize repulsions: Predicting Molecular Shapes: VSEPR If a molecule contains: • bonding pairs only – these angles are correct: triangular planar linear tetrahedral triangular bipyramidal octahedral Bonds and lone pairs determine shape. Use the notation AXnEm m lone pairs on central atom A n atoms bonded to central atom A Predicting Molecular Shapes: VSEPR Molecules may be described by their: • electron-pair (e- pair) geometry • molecular geometry (molecular shape) • These angle change (a little) if any “X” is replaced by a lone pair: • lone pair/lone pair repulsions are largest. • lone pair/bond pair are intermediate in strength. • bond/bond interactions are the smallest. Predicting Molecular Shapes: VSEPR AXnEm: 2 e- group central atoms (m + n = 2) Linear e- pair geometry 2 e- groups bond pairs These geometries may be different. lone pairs 2 0 AX2E0 1 AX1E1 linear 1 .. • Atoms can be “seen”, lone pairs are invisible. linear molecular geometry Predicting Molecular Shapes: VSEPR AX2E0 examples: Predicting Molecular Shapes: VSEPR AXnEm: 3 e- group central atoms (m + n = 3) 180.0° Cl Be Cl Triangular planar e- pair geometry Linear. 3 e- groups bond pairs “2” bonds, 0 lone pairs on C. (treat double bonds as 1 bond) Linear. O 3 0 AX3E0 triangular planar 2 1 AX2E1 .. 180.0° O C lone pairs angular (bent) H C C H Each H-C-C unit is linear. 1 2 AX1E2 .. 180.0° linear 180.0° molecular geometry 2 Predicting Molecular Shapes: VSEPR AX3E0 examples: Predicting Molecular Shapes: VSEPR AXnEm: 4 e- group central atoms (m + n = 4) 4 e- groups bond pairs 120° Cl B Cl lone pairs 4 Triangular planar. Tetrahedral e- pair geometry 0 AX4E0 tetrahedral Cl C C H H H Each C is AX3E0 = triangular planar. Predicting Molecular Shapes: VSEPR H AX4E0 All angles = tetrahedral angle. H C H AX3E1 triangular pyramidal .. H 1 .. 3 molecular geometry AX1E3? All molecules with only 1 bond are linear! 2 2 AX2E2 angular (bent) Predicting Molecular Shapes: VSEPR VSEPR applies to each atom in a molecule. • Alkanes: each C is tetrahedral. H AX3E1 Lone-pair/bond > bond/bond repulsion: H-N-H angle is reduced. H N H H AX2E2 Two lone pairs: H-O-H angle is even smaller. H O H Predicting Molecular Shapes: VSEPR Lactic acid: H Tetrahedral O Triangular planar C C C H H C .. O .. Central atoms with five or six e- pairs: Bond pairs Lone pairs Shape 5 0 Triangular bipyramidal 4 1 Seesaw 3 2 T-shaped 2 3 Linear .. .. .. .. H O H Expanded Octets Tetrahedral C Tetrahedral C Tetrahedral O O H 6 5 4 3 0 1 2 3 Octahedral Square pyramidal Square planar T-shaped • lone pairs repel the most. • they get as far apart as possible. 3 Expanded Octets Expanded Octets AXnEm: m + n = 5 Triangular bipyramidal e- pair geometry. Triangular bipyramidal Seesaw T-shaped F F F F P F F S F F Cl F F F F F Xe F Linear The atoms are non-equivalent. Green atoms are axial axial; blue atoms are equatorial equatorial. Expanded Octets Expanded Octets AXnEm: m + n = 6 Octahedral e- pair geometry: F All atoms are equivalent in AX6E0 F F S F F Octahedral Square pyramid Lewis dot + VSEPR predict molecular shapes, but… but How do atomic orbitals (s, p…) lead to these shapes? Valence bond theory: theory: bonds occur when partiallyoccupied atomic orbitals overlap. F Br F F Cl I Cl Cl HF – H(1s) overlaps F(2p) Valence Bond Theory This works for H2 and HF, but why does… • Be form compounds? • Be (1s2 2s2). • No unpaired e- to share. • Experiments show: linear BeH2, BeCl2, … • C form 4 bonds at tetrahedral angles? • • • • 74 pm Cl F Square planar Orbitals Consistent with Molecular Shapes H2 – H(1s) overlaps H(1s) F F C (1s2 2s2 2p2). 2px1 2py1 Two bonds? p orbitals are at 90° to each other Experiments show: tetrahedral CH4, CCl4, … 109 pm 4 Orbitals Consistent with Molecular Shapes Atomic orbitals (AOs) can be hybridized (mixed). sp Hybrid Orbitals Be compounds (BeH2, BeF2 …): • Sets of identical hybrid orbitals form identical bonds. • Number of hybrids formed = number of AOs mixed. One s orbital + one p orbital → two sp hybrids. Each sp hybrid (180° apart) holds one e-. Two equivalent covalent bonds form. sp2 Hybrid Orbitals B forms three sp2 hybrid orbitals: • One s orbital mixes with two p orbitals. • One p orbital remains unmixed. sp2 Hybrid Orbitals B compounds (BH3, BF3 …): Each sp2 hybrid (120° apart) holds one e-. Three equivalent covalent bonds form. sp3 Hybrid Orbitals C forms four sp3 hybrid orbitals: • One s orbital mixes with three p orbitals. • All p orbitals are mixed. sp3 Hybrid Orbitals N and O compounds (NH3, H2O…) have more e-: In C, each sp3 hybrid (109.5° apart) holds one e-. Four equivalent covalent bonds form. 5 sp3 Hybrid Orbitals Hybridization e- pair “Octet rule” molecules have tetrahedral shape. • sp3 hybridized (CH4, NH3, H2O, H2S, PH3, …) Head-to-head bond = a sigma bond (σ σ bond). bond There are: H • 4 σ bonds in CH4 σ bond • 3 σ bonds in NH3 • 2 σ bonds in H2O C H H Summary: Mixed s+p s+p+p s+p+p+p Hybrids (#) sp (2) sp2 (3) sp3 (4) Remaining Geometry p+p Linear p Triangular planar Tetrahedral d orbitals can also form hybrids: Mixed Hybrids (#) Remaining Geometry s+p+p+p+d sp3d (5) d+d+d+d Triangular bipyramid s+p+p+p+d+d sp3d2 (6) d+d+d Octahedral H Hybridization in Molecules with Multiple Bonds Carbon atoms form: • tetrahedral centers (CH4, CHF3 , C2H6…) = sp3 • triangular-planar centers (H2CO, C2H4 …) = sp2 H C C H H H Hybridization in Molecules with Multiple Bonds • a σ bond – head-to-head overlap of sp2 on each C atom. • a π bond – sideways overlap of p AOs on the C atoms. H C C H H Unhybridized C p orbitals each contain one e-. H C σ bond C The double bond in ethene is composed of: H C (sp2) + C (sp2) overlap (σ bond): H H C overlap H C H H Sideways overlap forms one π bond • the lobes above and below the plane together equal 1 bond Hybridization in Molecules with Multiple Bonds Hybridization in Molecules with Multiple Bonds Formaldehyde is similar: C also forms linear centers: • C2H2 (acetylene) = sp hybridized H C C H The triple bond is: • one σ bond • two π bonds • sp hybridization leaves two unmixed p orbitals on each C. 6 Hybridization in Molecules with Multiple Bonds Hybridization in Molecules with Multiple Bonds π bonds prevent bond rotation: σ bond: C (sp) + C (sp) overlap: H C C H Two p orbitals on each C contain a single e-. H C H C overlap H C C H Molecule ethane (CH3–CH3) ethene (CH2=CH2) ethyne (HC≡CH) C-C bonding σ σ+π σ+π+π C-C rotation yes no no Non-rotating double bonds allow cis-trans isomerism to occur. Two π bonds • above and below overlaps are 1 bond. • front and back overlaps are a second bond. Molecular Polarity Molecular Polarity • Most bonds are polar (e.g. C-O) • Water is polar (bond dipoles do not cancel) • O is δ-, C is δ+ (ENO = 3.5, ENC = 2.5) • But many molecules are nonpolar (e.g. CO2). O=C=O δ- 2δ+ O H arrow points to δ-, the + shows δ+ H + Net dipole Dipole, μ = 1.85 D δ- • The dipoles cancel because of CO2’s shape. • have equal size but point in opposite directions. Molecular Polarity Dipole moment (μ) is a measure of molecule polarity: Units: coulomb meter (Cm) Debye (D) weakly polar highly polar nonpolar (μ=0) Molecular Polarity Molecule H2 HF HCl HBr HI CH4 CH3Cl CH2Cl2 CHCl3 CCl4 μ (D) 0 1.78 1.07 0.79 0.38 0 1.92 1.60 1.04 0 A molecule is nonpolar if it is: • AXnE0 and all X are identical. CO2 AX2E0 linear CH4 AX4E0 tetrahedral CCl4 AX4E0 tetrahedral PF5 AX5E0 triangular bipyramidal • “divisible” into nonpolar AXnE0 shapes PCl3F2 triangular planar (PCl3) + linear (PF2 ) XeF4 linear (XeF2) + linear (XeF2 ) 7 Molecular Polarity AXnEm molecules are polar if they don’t divide into nonpolar shapes, and:: • m ≠ 0: H2O AX2E2 bent polar NH3 AX3E1 pyramidal polar H F + Molecular Polarity No net dipole C F C F F Net dipole F F F CHF3 is polar CF4 is non polar • The X in AXnE0 differ: CH2Cl2 PF4Cl AX4E0 AX5E0 tetrahedral polar triangular bipyramidal polar How polar? It depends on the number, type, and geometry of the polar bonds. Molecular Polarity Noncovalent Interactions Non polar AX5E0 and “X” differ. BUT divisible into nonpolar shapes: linear + triangular planar + PCl3F2 PCl5 Non polar AX5E0; identical X PCl4F Polar AX5E0 “X” differ Polar PF3Cl2 AX5E0 and “X” differ. Doesn’t divide into nonpolar shapes London Forces Also called dispersion forces. • Random e- motion produces a temporary dipole in one molecule which induces a dipole in another. ↔ 40 kJ/mol): Small molecule = few e- = weak attraction. Large molecule = many e- = stronger attraction. • Occur between all atoms and molecules. The only force between nonpolar molecules. Molecules attract each other. Intermolecular forces: • also called noncovalent interactions. • are small (compared to bonding forces). • do not include ionic or metallic-bonding forces. Three types: • London forces. • dipole-dipole attraction. • hydrogen bonding. London Forces Noble Gas # of ebp (°C) He 2 −269 F2 Halogen Hydrocarbon # of e- bp (°C) # of e- bp (°C) 18 −188 CH4 10 −161 Ne 10 −246 Cl2 34 −34 C2H6 18 −88 Ar 18 −186 Br2 70 +59 C3H8 26 −42 Kr 36 −152 I2 106 +184 C4H10 34 0 • Strength (0.05 More e- = larger attraction = higher b.p. 8 Dipole-Dipole Attractions Polar molecules attract each other. Strength: 5 ↔ 25 kJ/mol. Dipole-Dipole Attractions Nonpolar Molecules # of e- bp (°C) SiH4 18 −112 GeH4 36 −90 Br2 70 +59 Relative importance of dipole/dipole and London is hard to predict: HI HCl Hydrogen Bonding An especially large dipole-dipole attraction. • 10 ↔ 40 kJ/mol. • Occurs when H bonds directly to F, O or N. Polar Molecules # of e- bp (°C) PH3 18 −88 AsH3 36 −62 ICl 70 +97 Dipole small (0.38 D) large (1.07 D) London large (54 e-) small (18 e-) bp (°C) −36 −85 stickier Hydrogen Bonding H on one molecule interacts with O on another molecule. F, O & N are small with large electronegativities. • results in large δ+ and δ- values. H-bonds are usually drawn as dotted lines. Hydrogen Bonding Water is a liquid at room T (not a gas). 9