Enzymatic catalysis
advertisement

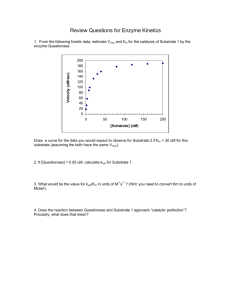
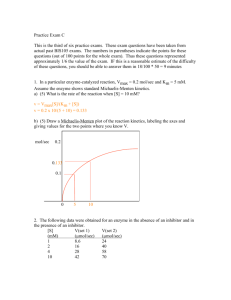
Lecture 10 Enzymatic catalysis Antoine van Oijen BCMP201 Spring 2008 Today’s lecture - Enzymes work by lowering ΔG‡ - Role of substrate binding - Role of catalytic groups - Enzyme kinetics: Michaelis-Menten - Inhibition mechanisms (Unless noted, figures from Lehninger; Principle of Biochemistry) 1 Enzymes speed up chemistry Nonenzymatic rate constant (knon in s-1) Enzymatic rate constant (kcat in s-1) Acceleration rate constant (kcat/knon) http://xray.bmc.uu.se/Courses/Tables/Tables.html From once every 100 million years (when the dinosaurs roamed the earth…) to 40 times a second! Enzymes lower activation energy ΔG‡ E+S ES EP E+P Enzymes do not change ΔG0, but lower ΔG‡ (remember, there’s a difference between energetics and kinetics) 2 Enzymes lower activation energy ΔG‡ Rate depends exponentially on activation energy! For decrease in ΔG‡ of 1 RT (≈ 2.4 kJ/mol): k = Ae(" #G m / RT) m kcat / kuncat = ! Ae("#Gcat / RT) Ae m ("#Guncat / RT) m =e m ["(#G cat "#Guncat ) / RT] = e1 $ 2.7 Reaction becomes 2.7 times faster ! For: 5 RT, reaction rate increases 10 RT, 15 RT, 25 RT, > 100 x > 20,000 x > 3 million x > 10 billion x Transition states, intermediates, and rate-limiting steps transition state intermediates Rate-limiting step is the transition with the highest ΔG‡ 3 Catalytic strategies 1) Noncovalent interactions between substrate and enzyme 2) Covalent interactions / chemical reactions between enzyme’s residues and substrate Binding provides major source of free energy ‘lock and key’ binding would be disadvantageous: Transition state needs to be stabilized, not substrate 4 Transition state analogs Transition-state analogs will bind tightly and inhibit catalysis Ester hydrolysis Carbonate hydrolysis How does binding help catalysis? Main energetic barriers contributing to ΔG‡: - Distortion Entropy Alignment w/ catalytic residues 5 Effect of entropy reduction on reaction rates Rate increase 1 105 M 108 M Role of catalytic groups 1) General acid-base catalysis 2) Covalent catalysis 3) Metal ion catalysis More on reaction mechanisms: Michael Wolfe next week 6 Enzyme kinetics: Michaelis-Menten equation Leonor Michaelis and Maud Menten (1913): Rate of catalysis by an enzyme is proportional to substrate concentration at low levels and becomes independent at high levels: E+S k1 k2 ES k-1 reaction substrate binding V0 = E+P [S] Vmax KM + [S] ! Enzyme kinetics: Michaelis-Menten equation E+S k1 k-1 k2 ES V0 = E+P [S] KM + [S] Fraction enzyme bound to ligand: Analogous ! to ligand binding, but k +k k KM = 2 "1 instead of "1 k1 k1 ! Vmax Vmax=k2[E]Total=kcat[E]Total [E]Total=[E]+[ES] kcat=turnover rate ! 7 Catalytic efficiency When [S] ! << KM : V0 = [S] [S] Vmax = kcat [E]Total KM + [S] KM + [S] V0 = kcat [S][E]Total KM 2nd order rate equation with units M-1s-1 ! Catalytic efficiency = kcat KM (theoretical upper limit ~ 109 M-1s-1) ! Some enzymes are diffusion-limited 8 Lineweaver-Burke representation V0 = ! [S] Vmax KM + [S] 1 KM + [S] KM 1 1 = = + V0 Vmax [S] Vmax [S] Vmax slope ! y-intercept (y=ax + b gives straight line; a=KM/Vmax. b=1/Vmax) Steady-state versus pre-steady state kinetics Steady state: [ES] is constant To gain information on initial steps to form E•S, pre-steady state techniques are needed 9 Pre-steady state techniques Stopped-flow / quenched-flow spectrophotometry Mix solutions at ~ 1 ms timescale and measure binding/activity Inhibition mechanisms Competitive inhibition Noncompetitive inhibition 10 Competitive inhibition Alcohol dehydrogenase Ethanol used as competitive inhibitor with methanol/ethylene glycol poisoning Noncompetitive inhibition HIV Reverse Transcriptase Nevirapine binds between polymerase and nuclease domains (Kohlstaedt et al., Science (1992); 256, 1783) 11 Competitive inhibition V0 = Where " = 1+ [S] Vmax "KM + [S] [I] KI and KI = [E][I] [EI] ! ! ! Competitive inhibitor changes KM (α•KM is ‘apparent’ KM) Noncompetitive inhibition V0 = Where # " = 1+ [S] Vmax KM + # "[S] [I] KI" and KI" = [ES][I] [ESI] ! ! V ! At high [I], v = max #" (enzyme ‘dilution’) ! 12 Kinetics test for determining inhibition mechanisms Lineweaver-Burke: 1 KM + [S] KM 1 1 = = + V0 Vmax [S] Vmax [S] Vmax slope y-intercept ! 1 KM + [S] "KM 1 1 = = + V0 Vmax [S] Vmax [S] Vmax Competitive: slope ! Noncompetitive: 1 KM + [S] KM 1 #" = = + V0 Vmax [S] Vmax [S] Vmax y-intercept ! Kinetics test for determining inhibition mechanisms 1 KM + [S] "KM 1 1 = = + V0 Vmax [S] Vmax [S] Vmax 1 KM + [S] KM 1 #" = = + V0 Vmax [S] Vmax [S] Vmax slope y-intercept ! ! 1 " 1% $ ' [S] # M & 1 " 1% $ ' [S] # M & ! Competitive ! Noncompetitive 13 Irreversible inhibition - Reactive substrate: Covalent binding to or destruction of essential residue (e.g., chymotrypsin +DIPF) - Suicide substrates: Substrate is converted into reactive species Sequential versus ping-pong 14 Enzyme classification http://www.chem.qmul.ac.uk/iubmb/enzyme/ Example: EC 1.1.1.27 Lactate dehydrogenase Single-molecule enzymology of β-Galactosidase Xie et al., Nature Chem.Biol. 2 (2006) 87 15 Concentration-dependence of waiting time Waiting time <τ> = 1/k Higher [S], shorter <τ> Michaelis-Menten from the single enzyme’s perspective ! ! Bulk-phase Lineweaver-Burke Single-molecule Lineweaver-Burke V0 [S] = kcat [E]T KM + [S] 1 [S] = kcat K " M + [S] [E]T KM + [S] KM 1 1 = = + V0 kcat [S] kcat [S] kcat " = ( [E]T=[E]+[ES], Vmax=k cat[E]T ) ! ! KM + [S] KM 1 1 = + kcat [S] kcat [S] kcat Same KM , same kcat 16 Ergodic theorem A measurement of some property of an ensemble at a given time should be equivalent to the long-time average of the same property on any one member Waiting time distributions E+S k1 k-1 ES kcat E+P Low concentration: Single-exponential decay At low [S], k1 is rate-limiting; at high [S], kcat is rate-limiting High concentration: multi-exponential decay: multiple kcat’s !!! 17 Enzymes are highly dynamic entities Enzymes do not have a constant kcat , but fluctuate over time Why is this happening? Rugged Energy Landscape: Distribution of conformations different enzymatic activities (kA, kB, kC) for different conformers A, B, C Distribution of barrier heights different transition rates (r AB, rAB, rAB) between different conformers A, B, C 18 Take-home lessons - Enzymes speed up chemistry by lowering ΔG‡ - Michaelis-Menten kinetics - Competitive and noncompetitive inhibition 19