DNA Cloning - Exploratorium
advertisement

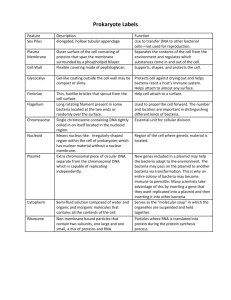
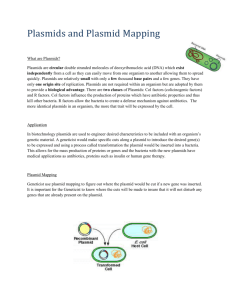
In the Beginning….There was Recombinant DNA Aka “Cloning” Using paper models and a modified “carnival” game, we’ll see how DNA from any species can be replicated and produced inside bacteria. Materials • Image of plasmid pTI2008; one or more per student • Page of four strands of DNA; one or more per student • Tape, one per student group • Scissors, one per student group • Large heavy paper clips, several per student • 50 ml beakers or other cups that do not easily tip; 8 per station, two stations per class To Do and Notice Preparing “Recombinant DNA” • Examine the plasmid, pTI2008. What features does it have? (A restriction site for EcoRI, an ampicillin resistance gene that confers ampicillin resistance to bacteria, a lac z gene whose product converts a substance in agar to a blue color, and “ori”, the origin of replication of the plasmid. • After an introduction to restriction enzymes, invite the students to cut their plasmid pTI2008 and their four DNA sequences at any EcoRI restriction site that they find. Remind them of the characteristic overhangs of the cut (see illo later in packet)/ • Examine all of the cut pieces and sticky ends. How many different ways could the various fragments re-connect (ligate) because of complementary sticky ends? • Make several different recombinant plasmids, using various sticky ended inserts and tape as the “ligase”. Also reanneal (seal) some plasmids without extra DNA. (a variation is listed at the end of the document). • Supercoil the plasmids by rolling them up tightly, and use a heavy paperclip to hold together. Transforming Bacteria Karen E. Kalumuck, Ph.D. Exploratorium Copyright 2008 • Split the class in half. For each half, create a station with 8-10 50ml beakers closely spaced and upside down. Have the students position themselves 7-8 meters from the beakers. • Shout “heat shock!” Invert the beakers to the upright position. Students should then toss each of their plasmids into the beakers, with the intent of having some enter the beaker “bacteria”. Allow several minutes for this. A long throwing distance is critical. When you shout “stop”, all activity must cease. Plating of the Bacteria • Remove the beakers and observe. Did all of the beaker bacteria receive a plasmid? Did some receive more than one? If you couldn’t see inside the beaker, would you be able to tell? How do scientists distinguish between the different possibilities? • Place the beaker bacteria on trays or areas covered with paper that says “Ampicillin”. You are plating the cells on agar that contains ampicillin. Which type of bacteria will grow here (ones with or without plasmids?) • How can scientists distinguish between bacteria that took up a plasmid with an insert, and those without? What’s Going On? There are literally hundreds of different restriction enzymes produced by bacteria as their own defenses against bacterial viruses and other invaders. In the early 1970’s scientists discovered that these enzymes cut all types of DNA, and that DNA from virtually every species could be introduced into a plasmid (a small circular extrachromosomal piece of DNA carried in bacteria). When plasmids are introduced into bacteria, the plasmids replicate within the bacteria and the bacteria will reproduce and pass the plasmids on to progeny cells, yielding millions of copies in just a few hours. In addition, the product of the gene inserted that was inserted into the plasmid can be produced by the bacteria, and can be detected with antibodies specific to the protein. Each restriction enzyme cuts a specific sequence of DNA. Overhanging (single stranded) sequences that are complementary can be made to join, and the enzyme DNA ligase will seal the ends. Bacteria are treated in the lab to allow DNA to adsorb to their surface (by chilling) then induced to bring the DNA into them with the heat shock (typically 49 degrees C). The heat shock period is very brief – Karen E. Kalumuck, Ph.D. Exploratorium Copyright 2008 if it lasted too long the cells will die, so the window of opportunity to transform bacteria with recombinant DNA is brief. The rate of transformation is very, very low – the carnival game of tossing the plasmids at a distance illustrates this. Many bacteria will not be transformed, but there are tricks to help in the identification. Plasmids have been engineered to help scientists conveniently identify bacteria that have been transformed with potential recombinant plasmids. The bacteria, after a recovery period at 37 degrees C in a nutrient broth, begin producing the products of genes on their plasmid. If a bacterium is transformed, it will be ampicillin resistant. It will grow on media impregnated with the antibiotic ampicillin. Bacteria that were not transformed will not grow. But how can you distinguish insert-containing bacteria from resealed plasmids? This plasmid is engineered to have the Lac Z gene, which will produce beta-galactosidease. This enzyme will degrade a chemical in the medium and turn the colony a blue color. The insertion site of the “foreign” DNA, the EcoR1 site, is within this gene. If the plasmid is recombinant (has some foreign DNA) the lac Z gene is nonfunctional, and the colony will appear white. What Next? The cloned plasmids may contain inserts different than what is desired. Their sizes and orientation are analyzed by restriction electrophoresis and/or PCR. The cDNA of interest can be checked to see if it makes a functional protein by growing up bacterial colonies containing the plasmid with DNA of interest, (which will make the protein, if the plasmid has a strong bacterial promoter before the insert). The cells are lysed, transferred to a replica plate, and mixed with radioactively labeled antibodies. Colonies producing the protein of interest will hybridize to the label, resulting in a black spot on X-ray film. So What? The cut-and-paste nature of restriction enzymes with DNA and the ability to produce millions of copies of any gene through “cloning” is the foundation of the biotech revolution. A few of the many important breakthroughts attributable to this basic technology include: Karen E. Kalumuck, Ph.D. Exploratorium Copyright 2008 • mass production of human medically significant substances, such as insulin and human growth factor • sequencing of the human genome – and every other genome. Variation for Creating Recombinant plasmids Provide each team of students two dye (dice). For each ligation event, allow the roll of the dice determine which event will take place. Each student should do at least 5 (or, your choice) ligations. All students at one lab table put their cut out fragments into a pile, and draw their insertions from this pile Roll of Dice Type of Ligation Event #2 2 plasmids ligate together # 3, 4, 5, 6, 7 Plasmid recircularizes without insert # 8, 9, 10 One copy of insert from pool into plasmid # 11, 12 Karen E. Kalumuck, Ph.D. Exploratorium Copyright 2008 Two copies of inserts from pool ligate together, and into single plasmid Randomized Ligation Events – Plasmids in Las Vegas!!! Groups at a table place all of their cDNA fragments and plasmids (both cut with EcoRI) into a single pile in the center of the table. Each individual will creat 5 ligated plasmids – with or without cDNA inserts as indicated by the following formula for the toss of two dice: Roll of Dice Type of Ligation Event #2 2 plasmids ligate together # 3, 4, 5, 6, 7 Plasmid recircularizes without insert # 8, 9, 10 into One copy of insert from pool plasmid # 11, 12 Karen E. Kalumuck, Ph.D. Exploratorium Copyright 2008 Two copies of inserts from pool ligate together, and into single plasmid pTI 2008 Karen E. Kalumuck, Ph.D. Exploratorium Copyright 2008 5’ 3’ ACTGGAATTCACATGCGAATTCGAACT TGACCTTAAGTGTACGCTTAAGCTTGA CGATGAAATTGGAGCATTCGAGACTTA GCTACTT TAACCTCGTAAGCTCTGAAT TGGCAGAATTCCGTATATCCTGAATTC ACCGTCT TAAGGCATATAGGACTTAAG GAATTCATGAATT CTAGCGCTAGCTAC CTTAAGTACT TAAGATCGCGATCGATG 5’ 3’ ACTGGAATTCACATGCGAATTCGAACT TGACCTTAAGTGTACGCTTAAGCTTGA CGATGAAATTGGAGCATTCGAGACTTA GCTACTT TAACCTCGTAAGCTCTGAAT TGGCAGAATTCCGTATATCCTGAATTC ACCGTCT TAAGGCATATAGGACTTAAG GAATTCATGAATT CTAGCGCTAGCTAC CTTAAGTACT TAAGATCGCGATCGATG Karen E. Kalumuck, Ph.D. Exploratorium Copyright 2008 Karen E. Kalumuck, Ph.D. Exploratorium Copyright 2008 Karen E. Kalumuck, Ph.D. Exploratorium Copyright 2008 “Sticky Ends” of EcoRI Restriction Endonuclease Cuts Karen E. Kalumuck, Ph.D. Exploratorium Copyright 2008