Biology
advertisement

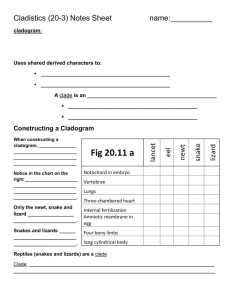
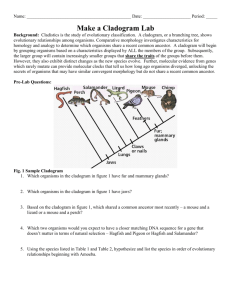
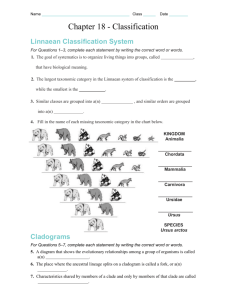
Biology Unit 6, Biodiversity/ Evolution, Lab Activity 6-2 In this lab1 you will develop a taxonomic classification and phylogenetic tree for a group of imaginary organisms called Caminalcules. The purpose of this lab is to illustrate the principles of classification and some of the processes of evolution. Taxonomists (people classifying organisms) use various methods to determine which taxa organisms should be placed in. The end product of classification is a cladogram. The ideal cladogram will reflect the evolutionary relationships between organisms. Various characteristics of an organism’s morphology (the structure of an organisms body) are used to determine relationships between organisms. Animals with four legs are probably more closely related to each other than to animals with flippers. Morphology can be misleading. Porpoise and tuna were once grouped together based on their similar morphology. Today we know that they are only distantly related. To help verify what morphology might suggest as a relationship researchers have turned more and more to genetic comparisons. By studying the genes and the proteins produced by those genes we can now get a very clear idea of how closely related a group of organisms are. You will use both morphology and genetics to determine a cladogram of the fictitious Caminals. We do these exercises with artificial organisms so that you will approach the task with no preconceived ideas as to how they should be classified. This means that you will have to deal with problems just as a taxonomist would. With real organisms you would probably already have a pretty good idea of how they should be classified and thus miss some of the benefit of the exercise. Research Question How does classification show relationships between animals? Protocol A ctivity 1 Classification Step 1 ) Cladistic Classification of Living Caminalcules based on Morphology In this step you will produce a cladogram of the living caminals on 11 x 17 inch paper. 1) Cut out and carefully examine the fourteen living species of Caminals and note the many similarities and differences between them. Use the following characteristics as an initial guide; Body Shape – Bulb/Long/Oval Eyes – None/1/2/Stalked Rear Legs – None/Single/Flipper/Toes Front Legs – None/Stub/Flipper/Tentacles/Toes Spotting – Shape/Number/Pattern/Size 1 Caminal figures from ‘Caminalcules’ Exercise by R. Gendron, Indiana University of Pennsylvania Lab6-2,Unit6,BiodiversityEvolution,ClassificationStudentLabPack2013 Greg Ballog 2013Page 1 of 9 2) Sort the specimens into like groupings based on body shape. This will give you three different clades of Caminals. We will call them the sluggers, bulb butts, and ovoids. 3) You will have 19 & 20 (sluggers) in one clade, 16, 1, & 24 (bulb butts) in a separate clade, and the remaining caminals in the third clade. By definition a clade is a group of organisms that share a common ancestor. It does not say who the ancestor was, just that they share one. It would be like two sisters being in a clade. Their mother would be their common ancestor even if they never knew their mother. 4) Now look at your three clades. Which two clades are most closely related? Sluggers Ovoids Big Butts Which two are most distantly related? Line them up across your table to show this relationship. 5) Figure 1 shows one way that the three clades can line up. You may not agree. That is fine but be prepared to explain your reasoning. You will notice that taken together the three clades can be considered a single clade since all of Figure 1 the caminals share a common ancestor. 6) Next sort each of the three clades into higher level clades based on one of the other characteristics. An easy clade to set apart is the group that includes 28, 14, & 13. None of them have eyes and would represent a smaller clade that is part of the ovoid clade. Your growing cladogram might look like the one shown in Figure 2. 7) Further sort each clade into higher and higher level clades until all of the caminals are separated. You will have a line running to each caminal when you are done. Ask your teacher to review your work before you glue the caminals to the 8 ½ x 14 inch piece of paper. Use a pencil to draw your lines in case you want to change it later. 8) Title your cladogram, "Living Caminal Cladogram." Put your name and class period on it. Include it with this lab report when you turn in your packet. Sluggers Ovoid eyeless Ovoid eyes Big Butts Figure 2 Answer Stop question #1-2 before continuing Lab6-2,Unit6,BiodiversityEvolution,ClassificationStudentLabPack2013 Greg Ballog 2013Page 2 of 9 EXTRA CREDIT: Make a mobile of your cladogram. A ctivity 2 – Phylogeny based on Molecular Evidence Step 2) Develop a cladogram based on Cytochrome c mutations 1) As a reminder of the process of translation (DNA→mRNA→Protein) use a mRNA code sheet to determine the amino acid sequence of the following DNA nucleotides. A code sheet may be found at http://en.wikipedia.org/wiki/Genetic_code Remember base pairing rules for DNA→mRNA are A to U, T to A, C to G, and G to C Amino Glycine Acid mRNA 5’G G C 5’ DNA 3’C C G C T A C A G A A G T T T C C T T T C5’ The sequence of nine amino acids you just decoded is the beginning of a much longer polypeptide that folds into the protein Cytochrome c. Read about Cytochrome c at http://en.wikipedia.org/wiki/Cytochrome_c Answer Stop question #3 before continuing The primary sequences of the 100 amino acids making up the Cytochrome c protein have been determined for a variety of organisms including all living caminals. The full sequence of each can be found on your course disk as either a pdf file, Excel file, or as a series of .fasta formatted files (more on .fasta files in a moment) or on the web on the Falcon Science site under Biology Web Downloads. Researchers have found that the sequence of amino acids in Cytochrome c varies among organisms due to periodic mutations in the DNA code. Since mutations accumulate over time it is possible to determine how closely related different species are by comparing their Cytochrome c sequences. Those with few Lab6-2,Unit6,BiodiversityEvolution,ClassificationStudentLabPack2013 Greg Ballog 2013Page 3 of 9 differences in the amino acid sequence had a recent common ancestor while those with a large number of differences diverged at a more distant time in the past. Answer Stop questions #4-6 before continuing 2) Compare all of the Caminal Cytochrome c sequences to determine patterns of descent. You can do this in one of two ways, manually or by using a web based sequence comparison tool. To compare the sequences manually (this is a lot of work-check out the SHORTCUT) you must; a) Print several copies of each sequence sheet and compare each Caminal with every other Caminal. b) Mark any differences. c) Count and record the differences in Data Table 1. d) Once you have completed all comparisons (91 total comparisons) you 6 17 26 can use the information to validate/update the cladogram you developed based on the morphology of the Caminals. A cladogram made from this data will use the number of differences in Cytochrome 2 2 c sequences between two Caminals as a measure of the distance 4 between them. (example; The analysis in Figure 3 shows that there were 4 amino acid differences between species 17 and species 26. This 2 provides a distance measurement for our cladogram . It is 4 mutations from Caminal 17 to Caminal 26. When we compare Caminal 6 to Caminal 26 we find 8 mutations have occurred .) Follow the link; Figure 3 < http://web.virginia.edu/Heidi/chapter5/Images/8883n05_29.jpg> To see a cladogram made from a Cytochrome c analysis. Answer Stop question #7 before continuing Ninety-one comparisons of 100 amino acid polypeptides is a lot of work. You can; a) do it yourself, b) split the work up among several of your classmates, or c) do it the easy way by using a computer based tool developed specifically to compare protein sequences. SHORTCUT Using BLAST to compare multiple amino acid sequences The U.S. government maintains NCBI (The National Center for Biotechnology Information) with web access open to anyone. The following instructions will walk you through using the NCBI website to compare your caminal sequences. You may also view the tutorial, ‘Comparing Protein Sequences using BLAST’, located on your course disk or on the Video Tutorials link of the Falcon Biology Home page. 1) Go to <http://blast.ncbi.nlm.nih.gov/Blast.cgi> to begin your analysis 2) Scroll down to <Basic BLAST> and select <protein blast> 3) Beneath the entry boxes on the new page click the <Align two or more sequences> box. A second set of data boxes will appear. 4) Click on the <Choose File> button below the first set of data boxes and navigate to where the .fasta files that contain the Cytochrome c sequences for the caminals is located. This may be on your course disk or at some other location if you downloaded the files from the school website. In both cases they are in a folder called <CaminalculeCytochromeSequences>. 5) Select the amino acid sequence file for the Caminal you want to analyze. After clicking on the file you will be returned to the BLAST search page. Lab6-2,Unit6,BiodiversityEvolution,ClassificationStudentLabPack2013 Greg Ballog 2013Page 4 of 9 6) Scroll down to the <Choose File> button in below the second set of data boxes and Navigate to, then select, the .fasta file, “Caminalcule AADatabase.” After clicking on the database file you will again be returned to the BLAST search page. 7) Scroll down to the button and select it. The selected sequence will be automatically compared to all of the sequences of the other living Caminals. Scroll down to the box titled, “Sequences producing significant alignments:”.In the, “Ident” is the number of matching amino acids. Subtract that number from 100 to obtain the number of differences. Record the differences in Data Table 1. Once you have compared all of the different Caminals you can have the BLAST sequencer produce a phylogenetic analysis of the data as follows; 9) At the top of any of your amino acid comparisons select [Distance tree of results]. A phylogenetic tree will be produced. Select <slanted> above the tree display to change the style of the tree. Use a screen capture tool to paste the cladogram into a word processor for printing. Turn your BLAST cladogram in along with the cladogram you produced by comparing caminal morphology. 10) Compare this BLAST cladogram with the Morphology cladogram you produced in Activity 1. 11) Produce a third cladogram that is a combination of the cladogram you produced based on morphology (Activity 1) and the one produced by comparing the Cytochrome c sequences (Activity 2). Include all three cladograms in your lab report packet. Data Table 1 Cytochrome c Analysis 1 2 3 4 9 12 1 2 3 4 9 12 13 14 16 19 20 22 24 28 13 14 16 Lab6-2,Unit6,BiodiversityEvolution,ClassificationStudentLabPack2013 19 20 22 24 28 Greg Ballog 2013Page 5 of 9 Stop Questions 1) What defines a clade? _____________________________________________________________ 2) What do the nodes of the cladogram represent? _________________________________________ 3) What function that is carried out by all cells is Cytochrome c involved in? (hint: we studied this process in unit 2) ___________________________________________________________________ 4) Changes in the order of amino acids are due to periodic mutations in the coding sequences of the protein. The first amino acid in the sequence you just decoded is glycine. What would happen if the RNA sequence that coded for glycine was mutated from GGC to GUC? ____________________________________________________________________________________ 5) What would happen if the mutation was from GGC to GGU? ________________________________ 6) Do all mutations cause a change in polypeptide sequences? ___________________________ 7) Using the example in figure 3, page 4; how many mutations are there between Caminal #6 and #17? ______ ? Is Caminal #6 more closely related to #17 or to #26? __________. Explain your reasoning (it is this explanation that will earn you credit). ___________________________________________________________________________________ Analysis Questions The analysis for this lab includes; 1) Your cladogram (11 x 17 sheet) based on Caminal morphology. 2) Your Cytochrome c Analysis Data Table 1 3) Your printed Blast Cytochrome c cladogram. 4) Your combined Morphological / Cytochrome c cladogram. Conclusion 1) 2) 3) 4) You have just used two methods to determine the phylogeny of a group of living organisms. They almost never agree. Explain why the two methods do not always agree. _______________________________________________________________ Which method do you have the most confidence in? __________________________ Explain your reasoning. _________________________________________________ ____________________________________________________________________ Why do you have more confidence in the method you chose (#2 above)? ___________________________________________________________________ Which method would you use to determine the relationships between living and fossil Caminals? __________________ Explain your reasoning ________________ ____________________________________________________________________ Lab6-2,Unit6,BiodiversityEvolution,ClassificationStudentLabPack2013 Greg Ballog 2013Page 6 of 9 Caminal Cytochrome c sequence Sheet #1 1 2 3 4 5 0 0 0 0 0 1 2 3 4 5 6 7 8 9 0 1 2 3 4 5 6 7 8 9 0 1 2 3 4 5 6 7 8 9 0 1 2 3 4 5 6 7 8 9 0 1 2 3 4 5 6 7 8 9 0 2 Z Z Z Z Z Z Z Z G D V F K G K K I S I M K C S H C K T E E K G G K Q V T G P N L H Y L F G R K T G Q 0 1 Z Z Z Z Z Z Z Z G D V E K G K P I S I M K G S Q C K T E E K G G K Q V T G P N L H G L F H R K K G Q 9 9 Z Z Z Z Z Z Z Z G D V C K G G K I S I M K C S P C H N V E K K F K H K C G P N L H G L F V WG T G Q 4 Z Z Z Z Z Z Z Z G D V E C N G K I S D M K C S P C H N E D K G C K H A C P P N L H G L R G L K V G Q 3 Z Z Z Z Z Z Z Z G D V E C N G K I S D M K C S P C H N E D K G C K H A C P P N L H G L R G L K V G Q 2 Z Z Z Z Z Z Z Z G D V E C N G K I S D M K C S P C H N E V K G G K H A C G P N L H G L F G L K V G Q 2 1 Z Z Z Z Z Z Z Z G D V E C G G K I S I M K C S P C H N E V K G H K H K C G P N L H G L F G L K V G Q 2 2 Z Z Z Z Z K Z Z G D V E K G G K I S I M L C S P C H N E E K G G K H K C G P N L H G L F G L K V G Q 1 Z T Z D M Z A Z G D V E K G G K I S I M K C S P A H N K E K G G K H K C G P N L H G L F G L K V G Q 6 2 Z Z Z D M Z A Z G D V E K G G K I S I M K C S P A H N K E K G G K H K C G P N L H G L F G L K V G Q 4 1 Z T Z D M Z Z Z G D V E K G G K I S I M K C S P C H N K E K Q G K H K C K P N L H G L F G L K V G Q 1 Z Z Z Z Z Z Z L G H V P L G N K I F I M K C S Q K I T V A K G G K H K T T P N L H G L F G S K T G Q 4 1 Z Z Z Z Z I Z Z G D V K L G K T I F I M K C M Q C H T V M P G G K H L Q G I N L H H L F G S K T G Q 3 2 Z Z Z P Z Z Z Z G D P Q L G K T I F I M K C M A S H T V E K G G H H I T G T N L H G P A V S K T G Q 8 Lab6-2,Unit6,BiodiversityEvolution,ClassificationStudentLabPack2013 Greg Ballog 2013Page 7 of 9 Caminal Cytochrome c sequence Sheet #2 6 7 8 9 0 0 0 0 0 0 1 2 3 4 5 6 7 8 9 0 1 2 3 4 5 6 7 8 9 0 1 2 3 4 5 6 7 8 9 0 1 2 3 4 5 6 7 8 9 0 1 2 3 4 5 6 7 8 9 0 2 S Q G Y S Y T A A N T V K G L I S G E D T L M E Y L E N P K K Y I WA T K M I F P M I K K N E R A D 0 1 A P G Y MY T A A N T V K G I K C G E D T L M E Y L E N P K K Y I P A C K M I F P M I K K N E R A D 9 9 A P G Y S Y T A A N MN K G Q Y WG E D T L M E Y L E N P K K Y N A D T K M I F V G I K K I E R A D 4 A P G Y R C P A A N K N H G I I WS E D T L M E Y L E N P K Q Y I P G K Q M I F A G N K K E E R A D 3 A P G Y R C P S A N K N H G I I WS E D T L M E Y L E N P K K Y I P G K Q M I F A G N K K E E R A D 2 A P G Y R C P Y A N K N H G I I WS E D T L M E Y L E N P K K Y I P G K Q M I F A G N K K E E R A D 2 1 A P G Y R Y P Y A N K N H G I I WS E D T L M E Y L E N P K K Y I P G K Q M I F A G N K K E E R A D 2 2 A P G Y R Y P Y A N K N H G I I WS E D T L M E Y L E N P K K Y I P G K Q M I F A G N K K E E R A D 1 A P WY S Y T A A N K N H G I I WS I D T L M E Y L E N P K K Y I P G T K Q I F A G N K K E E R A D 6 2 A P G Y S Y T A A N K N H G I I WS I D T L M E Y L E N P K K Y I P G T K Q I F A G N T K H E R A D 4 1 A P WY S Y T A A N K N H G I I WS E D T L M E Y L E N P K K Y I P G T K Q I F A G N K K E E R A D 1 A P G Y S Y T A A N K N K G I I WA E D T L M E Y L E N P K K A C E G T K M I F D WI K K E E R A D 4 1 A P G Y S Y T A A N K N K G I E WA E D T L M E Y L E N P K K Y S R L T K M I F V G M WK E E R A D 3 2 A P G Y S Y T A A N K N K G I I WA E D T L M E Y L E N P K K Y I P K T K M I F I G I K K L E R A D 8 Lab6-2,Unit6,BiodiversityEvolution,ClassificationStudentLabPack2013 Greg Ballog 2013Page 8 of 9 Lab6-2,Unit6,BiodiversityEvolution,ClassificationStudentLabPack2013 Greg Ballog 2013Page 9 of 9