biology 100- introductory biology
advertisement

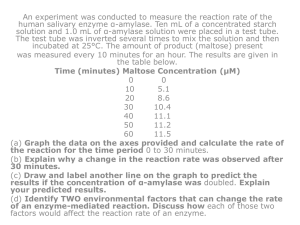
BIO 181 Enzyme Lab (pH) GENERAL INTRODUCTION: Alpha-amylase is a proteinic enzyme that hydrolyzes starch to produce maltose and other small sugars. Our saliva contains alpha-amylase (it is produced by the salivary glands), and so does our pancreatic juice (it is produced by the pancreatic secretory cells). Alpha-amylase is present in all organisms. We started this lab by extracting alphaamylase from three organisms: a thermophile bacteria (Bacillus licheniformis), a microscopic fungus (Aspergillus oryzae), and from the pancreas of a pig. Enzymes are affected by changes in pH and temperature. Each optimal has an optimal T and pH values at which they are more active. GENERAL OVERARCHING QUESTION: Can optimal T and pH values for enzyme activity be used to distinguish organisms? INTRODUCTION TO THE LAB: (Have students whiteboard table of variables before they start the lab) RESEARCH QUESTION: Does pH affect α-amylase activity (production of maltose)? HYPOTHESIS: pH has an effect on α-amylase activity. Optimal temperature values can be used to identify the source of different α-amylase. VARIABLES: (Have students whiteboard table of variables before they start the lab; three variables would be enough) Table of Variables (Enzyme lab, pH) Name I/D/C Symbol (Units) Description Temperature C T(°C) Test tubes @ 23°C Mass of maltose D m (g) Change in the amount of maltose produced Absorbance of maltose D Abs (Abs units) Change in absorbance values at 540nm pH I pH (pH units) Starch solution pH 5, 6, 7, 8, and 9 Substrate (starch) concentration C [c] (g/L) Keep pH the same in all tubes (1%) Volume of substrate (starch) C V (L) Keep pH the same in all tubes (1mL) α-amylase concentration C [c] (g/L) Keep pH the same in all tubes (XXX) Time of enzymatic reaction C t (s) Keep pH the same in all tubes (12 min) Wavelength C λ (nm) Keep pH the same in all tubes (540 nm) Pre-incubation time C t (s) Keep pH the same in all tubes (10 min) APPARATUS DRAWING: This should be a neat drawing that clearly illustrates all equipment and how each variable was measured. Draw and label!! PROCEDURE: The current protocol can be used as guideline. The procedure is the step-by-step process you go through to complete the experiment This should be what you actually did as opposed to what the directions said. You may be brief, but you should be thorough – don’t skip important steps!! DATA: pH (pH units) 5 6 7 8 9 Abs 540nm mg of maltose Table 1. Production of maltose at different pH for α-amylase A (B, or C) ANALYSIS: (Provide examples in Lab 1 as to how to calculate mg of maltose using the standard curve) Calculate mg of maltose using the equation that relates absorbance of maltose at 540 nm and mass of maltose (see maltose standard curve, Lab 1): Maltose Standard Curve absorbance at 540 nm 0.4 0.35 y = 0.1743x 0.3 0.25 0.2 0.15 0.1 0.05 0 0 0.5 1 1.5 2 2.5 m g of m altose Graph enzyme activity as measured by mg of maltose produced as a function of pH. Graph by hand using graph paper provided by your instructor (I have included Excel instructions just in case we decide they graph with Excel): Instructions for Graphing Using Excel 2007 Type your X-values (T or pH in our case) in column A Type your Y-values (mg of maltose in our case) in columns B, C, and D as shown above. Highlight values in all columns From the Insert menu, select “Chart” From “Chart Options” select “XY (Scatter).” Select first option “Scatter with data points connected by smoothed lines.” Click “Next.” Once in the new window, click “Series.” Name Series as alpha-amylase A, alpha-amylase B, and alpha-amylase C. Click “Next” Go to “Gridlines” and unselect “Major Gridlines.” Go to “Titles” and give a title to the two axis (X= Temperature (C) or pH; Y= mg of maltose). Title your chart as “Effect of Temperature on alpha-amylase Activity.” “Click Finish” Your graph is complete CONCLUSION: CLAIM: Make a claim that answers your research question. pH affects α-amylase activity (production of maltose). pH can be used to identify the source of our αamylase EVIDENCE: Supported by results. LAB SETTING: (Four datasets for each enzyme. Each table conducts two experiments) 2x Enzyme A 2x Enzyme C 2x Enzyme B 2x Enzyme B 2x Enzyme C 2x Enzyme A