Molecular Biology, Lecture 1
advertisement

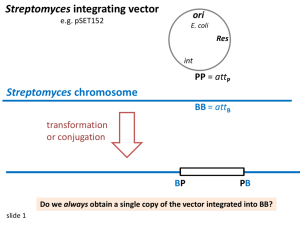
Molecular Biology, Lecture 1 A. Genotype / Phenotype Genotype: genes organism possesses (such as lacZ+) Phenotype: observable traits of an organism, (such as Lac+ grows on lactose) B. Mutation 1. Base substitution (same # of base pairs) if(codingRegion) * “missense” correct amino acid incorrect amino acid * “nonsense” correct amino acid premature stop codon TAG (amber) TAA (ochre) TGA (uped) 2. Insertions / Deletions (more / fewer base pairs) if(codingRegion) frame shift (3n+1) or (3n+2) 3. We no longer use the old term “point mutant” because it could be either substation/deletion/insertion C. Introducing foreign DNA into bacteria (a- in chromosome add a+) 1. Permanent Diploid a. Transformation: naked DNA moves outside to inside bacteria (a+ gene on plasmid) b. Transduction (a+ on DNA in transducent phage (virus) second copy of DNA from virus 2. Transient Diploid a. Conjugation / sexduction : A+ D. Complementation 1. Cell has a genetic element x (where x = gene or origin of replication or ….) X- cell is transformed with a X+ plasmid now have a diploid cell X+ / X- (genotype). What is the phenotype? X=gene : X+ phenotype, complementation occurred X=origin of replication: X- phenotype, complementation did NOT occur We cannot have the origin of replication on the plasmid and expect complementation (X+) to occur. Said again, that makes no sense – the plasmid is not going to give rise to X+ because the origin of replication where the transcription occurs is not in the cell’s DNA. (Note, convince yourself this is true) This is true for origin of replication, binding sites, promoter regions, etc. HOWEVER, transfactors (diffusible elements) will give rise to X+. They include: protein mRNA tRNA etc E. Mapping of Genetic Elements 1. Many techniques 2. 3 factor cross What is the gene order? Donor cell Recipient cell A+B+C+ A-B-C- If we want to know how we get/got A+, first we calculate how many of each cell: Permute: A+B+C+ A+B-C+ A+B+CA+B-C- 300 100 10 100 lowest frequency because it has highest # of recombination RULE: compare recipient progeny with the lowest # of recombinants to the parent who is most similar. A+B+C+ is the most similar, so C is the odd gene (“in the middle”). Gene order is ACB or BCA (note, there is no +/- written here) Recombinant DNA Techniques Review A. rDNA depds on bacterial proteins in the restriction / modification system 1. Restriction endonuclease & modification methlyase 2. the purpose is to recognize foreign DNA and destroy it 3. example ECOR1 (e.coli containing R plasmid , 1) 5’ GAATTC 3’ CTTAAG ECOR1 restriction endonuclease cleaves: o exonuclease recognizes ends o endonuclease recognizes middles DNA Replication Hemimethylated ECOR1 modification methylases (“overhanging end”, “sticky end”)