I. Plasmid Mediated Conjugation
advertisement
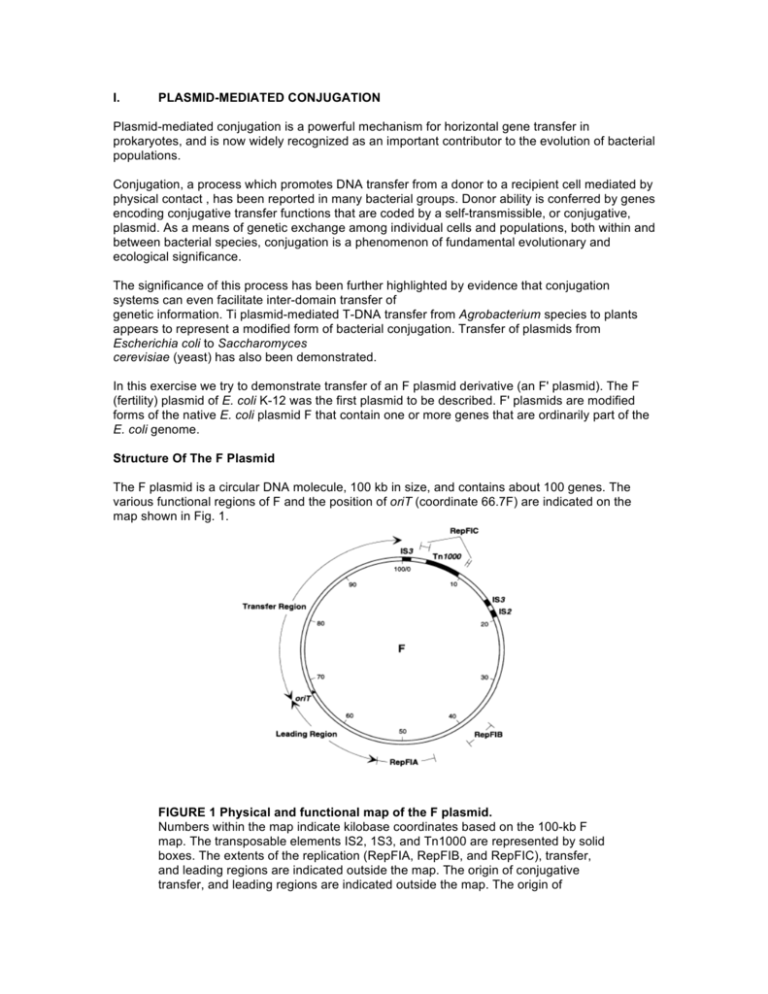
I. PLASMID-MEDIATED CONJUGATION Plasmid-mediated conjugation is a powerful mechanism for horizontal gene transfer in prokaryotes, and is now widely recognized as an important contributor to the evolution of bacterial populations. Conjugation, a process which promotes DNA transfer from a donor to a recipient cell mediated by physical contact , has been reported in many bacterial groups. Donor ability is conferred by genes encoding conjugative transfer functions that are coded by a self-transmissible, or conjugative, plasmid. As a means of genetic exchange among individual cells and populations, both within and between bacterial species, conjugation is a phenomenon of fundamental evolutionary and ecological significance. The significance of this process has been further highlighted by evidence that conjugation systems can even facilitate inter-domain transfer of genetic information. Ti plasmid-mediated T-DNA transfer from Agrobacterium species to plants appears to represent a modified form of bacterial conjugation. Transfer of plasmids from Escherichia coli to Saccharomyces cerevisiae (yeast) has also been demonstrated. In this exercise we try to demonstrate transfer of an F plasmid derivative (an F' plasmid). The F (fertility) plasmid of E. coli K-12 was the first plasmid to be described. F' plasmids are modified forms of the native E. coli plasmid F that contain one or more genes that are ordinarily part of the E. coli genome. Structure Of The F Plasmid The F plasmid is a circular DNA molecule, 100 kb in size, and contains about 100 genes. The various functional regions of F and the position of oriT (coordinate 66.7F) are indicated on the map shown in Fig. 1. FIGURE 1 Physical and functional map of the F plasmid. Numbers within the map indicate kilobase coordinates based on the 100-kb F map. The transposable elements IS2, 1S3, and Tn1000 are represented by solid boxes. The extents of the replication (RepFIA, RepFIB, and RepFIC), transfer, and leading regions are indicated outside the map. The origin of conjugative transfer, and leading regions are indicated outside the map. The origin of conjugative transfer (oriT) is denoted by a triangle indicating the direction of single-stranded DNA transfer (leading region transferred first). Leading Region The F DNA sequences located between the origin of conjugal transfer, oriT, and RepFIA are presumed to be the first to enter the recipient cell during conjugation and have therefore been designated the leading region. Leading-region gene products are thought to assist in establishing F DNA in the recipient. Autonomous Replication The RepFIA region is responsible for the typical replication of F in the E. coli host. Replication and associated maintenance and partitioning mechanisms act in concert to maintain the plasmid at one to two copies per cell. The secondary replication region, RepFIB, is independently functional and can sustain plasmid replication in the absence of RepFIA. The RepFIC region includes an incomplete remnant of a replication system that is used by some other related plasmids. Transposable Elements The F sequence includes a single copy of Tn1000 and IS2 and two copies of IS3. Insertional inactivation of the transfer region regulatory gene, finO, by IS3 is fortuitously responsible for the constitutively high levels of conjugative transfer exhibited by F. These elements also serve as the substrate for homologous recombination events with the host cell genome leading the genome fusion (Hfr strains). F' plasmids are generated by subsequent imprecise excision of F from the composite genome of an Hfr strain. Conjugative Transfer Including the oriT site (map position 66.7F), the transfer (tra) region encodes all of the F genes known to be required for efficient conjugative transfer. Figure 2. Figure 2 depicts the stages of intercellular contact and DNA transfer thought to occur during Fmediated conjugation. These are as follows: Contact between F+ donor and F– recipient cells is initiated by interaction between the tip of an F pilus and the recipient cell surface. Susequently, depolymerization of the pilus subunits draws the cells into close physical contact. Conjugating cells are typically aggregated in close, stable wall-wall association. A large number of the F products required for conjugative transfer are involved in F-pilus synthesis and aggregate stabilization. A protein complex (oriT complex) assembles on the origin of transfer. One DNA strand in the oriT site is “nicked” by an endonuclease (TraI) that also catalyzes the covalent attachment of the 5' end of the nicked strand DNA to the protein. TraI, is also a helicase and can unwind the nicked strand in the 5' to 3' direction. During transfer, this protein is associated with the site of intercellular connection between donor and recipients cells, and through which this single strand of DNA is passed. TraI may also subsequently catalyze re-circularization of the transferred strand to terminate transfer Replacement strand synthesis in the donor and complementary-strand synthesis in the recipient depend on host cell enzymes. Figure 2 shows DNA synthesis in the donor by a rolling-circle mechanism.









