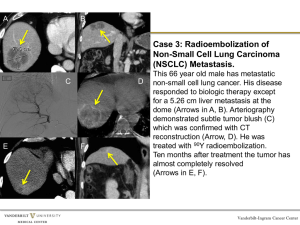
Pharmacogenetic and Germline Prognostic Markers of Lung Cancer
advertisement

Pharmacogenetic and Germline Prognostic Markers of Lung Cancer Anne M. Horgan, MB BCh, MRCPI; Boming Yang; Abul Kalam Azad, MBBS, MSc, PhD; Eitan Amir, MB ChB; Thomas John, MBBS, PhD, FRACP; David W. Cescon, MD; Paul Wheatley-Price, MB BCh; Rayjean J. Hung, PhD, MSc; Frances A. Shepherd, MD, FRCP(C); Geoffrey Liu, MD, MSc, FRCPC Princess Margaret Hospital, 610 University Avenue, Toronto, Ontario, Canada, M5G 2M9 1 Supplementary Table 1A. Pathways/Polymorphisms most frequently studied Genetic Polymorphism XRCC1 4-13 XRCC1 Arg399Gln Studies Comment: Positive Findings 10 studies 4 Asian; 6 Caucasian n= 38-229 XRCC1 Arg194Trp 4 studies 3 Asian; 1 Caucasian n=36-229 XRCC1 Arg280His 1 study Asian; n=229 TREATMENT WITH PLATINUM Gln allele → better OS in three Caucasian studies 4-6 Gln/Gln → worse OS in one US study 9 Gln/- → worse grade 3 or 4 toxicity in a single Asian study [AOR 2.05 (p=0.04)] 13. Arg/Trp → better RR to platinum in single study (p=0.04) 7. Trp/- → worse toxicity with gemcitabine/docetaxel (p=0.03) 10 Arg/Arg → better PFS (radiation alone) (p=0.03) 11 Arg/Arg → no association with cisplatin in single study 13. No significant associations 11 ERCC15,6,20-26,75,82 ERCC1 C118T DNA REPAIR ERCC1 C8092A ERCC1(G262T;T433C; C4855T) XPD 4,5,8-10,21,25,83,84 XPD Asp312Asn 9 studies 4 Asian; 5 Caucasian n=65-245 6 studies 2 Asian; 4 Caucasian n= 119-423 1 study Asian; n=162 TREATMENT WITH PLATINUM C/C → better OS/RR in three Asian studies20-22 C/C → No associations in 4 Caucasian studies6,24-26 C/- → independently associated with better response in a single European study compared to T/T [OR 0.10; p=0.03] 5 C/C → better OS in 2 studies 23,26. C/C → poorer OS in 2 studies 5,75. C/C → less toxicity in a single study 24. 262T/T → worse OS [AHR 1.98; p=0.02] :SCLC only, treated with carboplatin/VP16 82 6 studies 2 Asian; 4 Caucasian n=39-209 7 studies 2 Asian; 5 Caucasian n=39-248 1 Study Asian; n=209 TREATMENT WITH PLATINUM 312Asn/Asn →worse OS in a single study (p=0.003) 9. No significant associations in remaining studies 5,21,25,83,84 751Lys/Lys → higher incidence Gr 4 neutropenia (p=0.02) 10. No significant associations in remaining studies 4,5,8,21,25,83 6 studies 1 Mixed; 2 Caucasian 3 Asian n=53-214 NO GEMCITABINE 37C/C - 524T/T haplotype → better DFS compared to 37A/C524C/T [AHR 0.59; p= 0.05] 85 524C/T → better RR than 524C/C or T/T with platinum therapy (p=0.05) 86 TREATMENT WITH GEMCITABINE 37A/C-524C/T haplotype → better RR compared to other allelotypes (p=0.04) 88 No association in the overall population in a second study 6 Pathway analyses 75,76 25 SNPs in 16 genes in DNA repair pathways 1 Study Caucasian; n=229 6 polymorphisms in nucleotide excision gene associated with OS, though not significant 75 109 SNPs in 50 DNA repair genes 1 Study Caucasian; n=700 15 polymorphisms associated with OS mostly in nucleotide excision and base excision repair pathways; interactions with platinum76 XPD Lys751Gln XPD C156A XPD Asp711Asp RRM1 6,85-89 RRM1 -A37C RRM1-C524T RRM1 T756C RRM1 C269A 156A/A → higher incidence of grade 3/4 haematological toxicity in platinum-treated patients than C/C or C/A (p=0.01)84 2 DNA REPAIR Supplementary Table 1A cont’d. Pathways/Polymorphisms most frequently studied Genetic Polymorphism Others 7,22,25,73,87,89-92 iASPP A67T ATM A60G APE1 Asn148Glu MDR1 (C3435T; 2677GT/3435CT*) CDA Lys27Gln CDA C435T hMSH2 gIVS12-6T/C EGFR TKI XPG (His46His; His1104Asp; XPA A23G BRCA1 Tagging polymorphisms EGFR 32-39,93-95 EGFR intron 1 (CA)nS/L (S=short ; L=long) Comment: Positive Findings 1 Study Asian; n=230 iASPP 67A/- → better RR (p=0.01) only in a cisplatin/radiation subgroup 22 2 Studies 1 Asian; 1 Caucas n=54-62 2 Studies 1 Asian, 1 Caucas n=53, 65 1 Study Asian; n=156 2 Studies Asian; n=82-115 3435C/C → better RR in Asian study compared to T/(p=0.03) 92. No association in Caucasian study 87 1 Study Asian; n=300 7 studies 4 Asian; 3 Caucasian n = 70-170 EGFR -G216T EGFR -216G/-191C* 3 studies 3 Caucasian n= 92-173 EGFR G2607A EGFR C2982T 3 Studies Asian n=46 - 84 ABCG2 C421A ABCG2 G34A; ABCC3 C211T; ABCC3 G3890A; ABCC3 C3942T; 2 studies Caucasian; n=173,349 p53 / MDM2 43-50 p53 Arg72Pro p53 Intron 3 (single study) MDM2 T309G OTHER Studies GST 5,52,53,63,65,66,96-99 GSTM1-null/present GSTT1-null/present GSTP1 Ile105Val GSTP1 A/G GSTP1Ala114Val γ-GCS-7/8 CDA Lys/Lys → better OS in a single study (p=0.002) compared to Lys/Gln or Gln/Gln 25 hMSH2 T/T →worse OS (p=0.003) in a single study compared to C/C or T/C 91. XPA G/- → better response than AA to platinum-based therapy [OR 0.2; p=0.004] 90 No association in second study 7 AACC wild type haplotype → shorter survival [AHR 2.10; p=0.001] 73 TREATMENT WITH GEFITINIB LL → worse OS in a single Asian study (p=0.04) 32 L/- → worse PFS in a Caucasian study [AHR 1.9; p=0.03] 35 LL → worse RR in an Asian study alone or with a D994D polymorphism (p<0.001) 37 L → worse RR (p=0.03) and TTP (p=0.01) in Asian study 36 NO GEFITINIB L → better OS in a single US study [HR 0.66; p=0.03] 38 TREATMENT WITH GEFITINIB 216G/G → worse PFS, alone or in combination with Intron 1 L allele in one study 35 EGFR*1(-216G/-191C) haplotype → worse OS (p=0.02), only in ECOG PS 0 /1 patients, in 2nd study 33 TREATMENT WITH GEFITINIB 2607G/G → better OS than A/- (p=0.07)94 2982 C/C → longer PFS compared to T/- (p=0.002)37 TREATMENT WITH GEFITINIB ABCG2 421A/- →.higher incidence of diarrhea (p=0.005) 39 NO GEFITINIB ABCG2 421A/- → worse OS than C/C [HR 1.6 (1.04-2.47)] (with platinum) 93 ABCC3 211T/- variants → poorer PFS [HR 1.8 (1.13-2.82)] in a small cell subgroup 93 7 studies 3 Asian; 4 Caucasian n=101-619 2 studies 1 Asian; 1 Caucasian n=148, 383 Arg/Pro → worse OS in 1 study [AHR 2.3; p=0.02] 43 Pro/Pro → worse OS in 1study 44 and worse PFS in a 2nd46 Pro/Pro → worse OS on subgroup analysis in a third study45 309G/G → worse OS in early stage [AHR 1.6; p=0.04] 50 309G/- → worse OS in advanced disease [AHR 1.7;p=0.03 46 10 studies 4 Asian 6 Caucasian n=81-425 GSTM1-null → shorter OS in two studies [RR of death 1.36] 52 & [AHR 4.1] 53 GSTT1-null → shorter OS in a single study [AHR 2.1; p=0.01] 98. Similar trend in 2nd study but small numbers (n=6) 5 GSTP1 114 Val/- → longer survival in one study [AHR 0.75; p=0.037] compared to Ala114Ala 97 3 Supplementary Table 2A. Additional polymorphisms assessed in multiple studies Pathway/Gene Matrix Metalloproteinase 54,55,100 MMP1 1607 1G/2G ; MMP2 (C1306T, C735T ) ; MMP3 (1171 5A/6A, A706G, Glu45Lys) ; MMP7 A181G MMP9 (C1562T, Pro574Arg, Arg279Gln, Arg668Gln) MMP12 (A79G; A1082G) FGFR 101,102 FGFR4 Gly388Arg Studies Comment: Positive Findings 3 studies 1 Asian; 2 Caucasian n=90 - 561 Two large studies assessed multiple SNPs 54,55. MMP12 1082G/- alleles → worse OS in stage I NSCLC [AHR 1.94; p=0.002] compared to A/A 54 MMP9 279 Gln/Gln → worse OS compared to Arg/- [HR 1.6; p=0.023] in a large Asian study 55 MMP2 -735C/C → worse prognosis than T/- alleles in a smaller study [RR of death 2.6; p=0.05] 100 2 studies Caucasian n=274, 619 Arg/- → poorer OS in a single study (adenocarcinoma) [HR 1.6; p=0.008] compared to Gly/Gly 102; Arg/- → no association in the larger study (SCLC+NSCLC) Irinotecan Metabolism 56,57,103-105 UGT1A7 (*1,*2,*3,*4) 3 studies UGT1A1 (*6,*28,*60) 2 Asian; UGT1A16/7; UGT1A9*22 1 Caucasian n=47-118 ABCB1 (G2677T/A, C1236T, 1 study C3435T); ABCC2 (-C24T, Asian; n=107 G1249A, C3972T) SLCO1B1 G11187A SLCO1B1 1 study A388G SLCO1B1 T521C Asian; n=81 Vitamin D Receptor58,59 VDR Cdx-2 G/A 2 studies VDR Fokl C/T Caucasian VDR Bsml C/T n=294, 373 Folate Metabolism 60,61,106,107 MTHFR (Ala222Val, Arg594Gln) MTHFS Thr202Ala MTRR Ser175Leu (14 SNPS genotyped) MTHFR C677T MTHFR A1289C TS VNTR H/- vs L/L VEGF 62 VEGF (G405C, C936T, T460C) L-myc 63,64,108-111 L-myc Ser362Thr L-mycEcoR1 RFLP CYP 65-67 CYP2E1; CYP1A1 CYP2D6*4; CYP3AP1*3; CYP3A5*3; CYP2C19*2 CYP2C19*3; CYP2D6*2 1 study Caucasian n=619 3 studies 1 Asian 2 Caucasian n=127 - 295 UGT1A1*6 /6→ shorter PFS (p=0.001) and OS (p=0.17) in a single Asian study (irinotecan treated) 56 No association with other SNPs and survival OR response in irinotecan treated patients103,104. ABCC2 24T/T and 3972T/T → higher RR (p= 0.31 and 0.05) and longer PFS (p=0.01 and 0.02) than ABCC2 24 C/- and 3972 C/- 57 SLCO1B1 521C/- → higher incidence of Grade 4 neutropenia (p=0.008) 105. No association with response. Early stage (Squamous Cell):VDR Cdx-2 A/- → better survival [AHR 0.56; p=0.05] Combined “protective” genotypes and A-C-T haplotype better OS 59 Advanced stage: VDR Fokl T/-, and G-T-C haplotype → worse survival 58 Val222Val → shorter OS in SCLC only [HR 1.92 (1.03.58)] compared to Ala222Ala Arg594Gln → longer OS in overall population [HR 0.68 (0.46-1.0)] compared to Arg594Arg. Thr202Ala and Ser175Leu variant carriers → shorter OS in NSCLC 60 C677T → no association with OS in two studies 106,107 T677T → better PFS than C/T or C/C on univariate analysis in a third study (p=0.03) 61 TS L-group →longer OS only in combination with stage I and MTHFR C-group (p=0.03) 107. 1 study Caucas; n=462 VEGF 405 C/- → better 5year OS [AHR 0.70; p=0.008]. 6 studies 3 Asian; 3 Caucas n=83-252 L-mycEcoR1 RFLP S allele → shorter OS in one Asian study64, longer OS in a second Asian study 63and no association in two Caucasian studies 108,111 2 studies Mixed; n=87 Asian; n=232 1 study Asian; n=59 CYP2E1 wild type → better 5 year survival (p=0.02) 66 CYP1A1 non-susceptible homozygous → better OS in advanced NSCLC (p=0.005)65 Treatment with Vinorelbine 67 CYP2D6*4 C/C (p=0.02) and CYP3AP1 A/A → better RR (p=0.004) CYP3A4*53 G/G → worse RR (p=0.004) 4 Supplementary Table 3A. Polymorphisms assessed in single studies Pathway/Gene TRIT1110 TRIT1 Phe202Leu Inflammatory 112 TGF-β codon (10T/C, G25C); IL-6 G174C; IL-10 (G1082A, C592A,C819T); IFN-γ T874A; TNFα G308A IGF 113 3’UTR IGF2R-A2/B2 Immune 114 MBL2 Y/X -289G>C; MBL L/H -618G>G; MBL A/D Exl -34C>T; MBL A/B Ex 127G>A; MBL A/C Ex1 -18G>A Others 61,89,93,115-120 KRAS2 Rsal A1/A2; PTHLH VNTR; CDKN1B Val/Gly; M4(BstXI; StuI) LRMP (Val141Leu; Ser197Cys) CASC1 (rs12367971, rs10842496, rs10842501,rs10842502) Polysomy 7 MUC1 VNTR LS PDCD5 rs1862214 C/G FAS G1377A; FAS A670G; FASL C844T DCK C3122T; DCTD T315C; POLA2 G1747A; S28A1 (419 Ins/Del, G565A, C709A, G1368A, C1528T, G1561A); S28A2 (C65T, C225A); S28A3 A338G; TYMS (1002R/3R, G58C, 15705Ins/Del) GGH C452T; SLC 19A1 G80A; TS 5UTR T/R; TS 5UTR 3C/G CCND1 A870G CDKN1A (p21 codon) 31Ser31Arg Studies Comment : Positive Findings 1(+ replication ) Predominantly Caucasian n=246,335 Leu allele → worse OS in Italian group (adenocarcinoma) [HR 1.7; p=0.04)]. Leu allele → better OS in Norway group: [HR 0.5; p=0.02]. 1 study Caucasian; n=44 TGF-β T/T → better OS compared to C/C (p=0.01). 1 study Caucas; n=103 A2/B2 → worse OS (p=0.05) and +ve synergistic effect with p53 inactivation. 1 study Mixed; n=731 MBL2 X/- → improved survival in Caucasians, p=0.001. MBL2 LXA haplotype → improved survival. 1 study Caucas; n=213 1 study Caucasian n=361 1 study Caucas; n=82 1 study Asian; n=56 1 study Caucas; n=254 1 Study Asian; n=338 1 Study Asian; n=53 KRAS2 A2/- → better survival (p=0.008). PTHLH allele2 → worse survival (p=0.03) 116 LRMP 141 Val/Val → better OS in <65years (p=0.005) 117 No association with OS in overall population. Treatment with Gefitinib: High Polysomy 7 → better OS (p=0.004) 115 LL → worse OS only in adenocarcinoma compared to S/- (p=0.11) 118 G/- → worse survival [HR 1.8, p=0.003) compared to C/C 120 FAS 670G/- → worse OS than AA [HR 1.5, p=0.047] 119 S28A2 65 T/- → independently associated with improved OS [HR 0.3, p=0.002] 89 1 Study Asian; n=127 1 Study Caucas; n=244 1 Study Mixed; n=155 No significant associations with PFS or toxicity 61 GG → better response to platinum compared to A/- (p=0.04). No association with survival 121 Ser/Ser shorter survival compared to Ser/Arg or Arg/Arg (p=0.097) 122 Abbreviations Tables1A-3A: Caucas: Caucasian; DFS: disease-free survival; AHR: adjusted hazard ratio; OS: overall survival; RR: response rate; GI: gastrointestinal; gem: gemcitabine; tox: toxicity; PFS: progression-free survival; ECOG PS: European Cooperative Oncology Group Performance Status; SCC: squamous cell carcinoma; NSCLC: non-small cell lung cancer; SCLC: small cell lung cancer. *Haplotype. References 1-81 are found in the main text and 82-122 are found in the supplemental text as supplementary references. Supplementary Tables 1B,2B,3B show the RefSNP identifiers of the corresponding polymorphisms in Tables 1A,2A,3A. 5 Supplementary Table 1B. Sequence variants and their RefSNP corresponding to Supplementary Table 1A Gene SNP DNA Synthesis / Repair XRCC1 Arg399Gln Arg194Trp Arg280His RefSNP ERCC1 C118T C8092A G262T T433C C4855T rs11615 rs3212986 rs2298881 rs3212930 rs3212961 XPD Asp312Asn Lys751Gln C156A Asp711Asp rs1799793 rs13181 rs238406 rs1052555 RRM1 -A37C -C524T T756C C269A A67T A60G Asn148Glu N/A N/A N/A N/A rs6966 rs664143 rs1130409 3435C/T G2677T Lys27Gln A79C C435T gIVS12-6T/C A23G His46His His1104Asp rs1045642 rs2032582 rs2072671 rs2072671 rs1048977 rs2303428 rs1800975 rs1047768 rs17655 1(CA)nS/L -G216T -A191C D994D C2982T G2607A C421A G34A N/A rs712829 rs712830 rs2293347 rs2293347 rs1050171 rs2231142 rs2231137 C211T G3890A C3942T G565A N/A rs11568591 rs2277624 rs2290272 iASPP ATM APE1 MDR CDA hMSH2 XPA XPG EGFR EGFR ABCG2 ABCC3 CNT1 rs25487 rs1799782 rs25489 Gene p53 P53 SNP RefSNP Arg72Pro Intron 3 rs1042522 N/A MDM2 MDM2 T309G Glutathione S-transferase GSTP1 Ile105Val A/G Ala114Val γ-GCS 7/8 rs2279744 rs1695 N/A rs1138272 N/A N/A: not available 6 Supplementary Table 2B. Sequence variants and their RefSNP corresponding to Supplementary Table 2A Gene SNP Matrix Metalloproteinase MMP1 1607 1G/2G MMP2 C1306T C735T RefSNP MMP3 1171 5A/6A A706G Glu45Lys rs3025058 N/A rs679620 A181G C1562T Pro574Arg Arg279Gln rs11568818 rs3918242 rs2250889 rs17576 Arg668Gln A79G A1082G rs17577 N/A rs652438 Ser362Thr EcoRI RFLP rs3134614 N/A MMP7 MMP9 MMP12 L-myc L-myc FGFR FGFR4 Gly388Arg Irinotecan Metabolism UGT1A1 *6 *28 *60 6/7 UGT1A7 *1 *2 *3 *4 UGT1A9 *22 ABCB1 T2677A C1236T C3435T rs1799750 rs243865 rs2285052 rs351855 Gene Vitamin D Receptor VDR SNP RefSNP Cdx-2G/A Fokl C/T Bsml C/T N/A N/A N/A Ala222Val Arg594Gln A1289C rs1801133 rs2274976 rs1801131 Ser175Leu Thr202Ala VNTR H/L/L rs1532268 rs8923 N/A N/A N/A *4 *2 N/A N/A rs3892097 rs16947 CYP2C19 *2 *3 rs4244285 rs4986893 CYP3A5 CYP3AP1 *3 *3 rs776746 N/A Folate Metabolism MTHFR MTRR MTHFS TS CYP CYP2E1 CYP1A1 CYP2D6 rs4148323 N/A N/A N/A N/A N/A N/A N/A N/A rs2032582 rs1128503 rs10456642 ABCC2 -C24T G1249A C3972T rs717620 rs2273697 rs3740066 SLCO1B1 G11187A A388G T521C rs4149015 rs2306283 rs4149056 N/A: not available 7 Supplementary Table 3B. Sequence variants and their RefSNP corresponding to Supplementary Table 3A Gene TRIT1 TRIT1 VEGF VEGF Inflammatory TGF-b IL-6 IL-10 TNFa IFN-γ IGF IGF2R Immune MBL2 MBL Others KRAS2 PTHLH CDKN1B M4 LRMP CASC1 Chr7 MUCI SNP RefSNP Phe202Leu rs3738671 Gene Others FAS G405C C936T T460C rs2010963 rs3025039 rs833061 FASL NQO1 STK15 codon 10T/C codon 25G/C G174C G1082A C592A 819C/T G308A T874A rs1800470 rs1800471 rs1800795 rs1800896 rs1800872 rs1800871 rs1800629 rs2430561 PDCD5 CDA 3'UTR A2/B2 N/A Y/X -289G>C L/H -618G>G A/D ExI -34C>T A/B ExI -27G>A A/C Exl -18G>A rs7096206 rs11003125 rs5030737 rs1800450 rs1800451 RsaI A1/A2 VNTR Val/Gly BstXI, StuI Val141Leu Ser197Cys R33S Intronic Intronic Intronic Polysomy 7 VNTR LS N/A N/A N/A N/A rs7969931 rs1908946 rs10842496 rs12367971 rs10842501 rs10842502 N/A N/A DCK DCTD POLA2 S28A1 S28A2 S28A3 TYMS GGH SLC19A1 TS ABCC3 RAD23 CCND1 CDKN1A N/A: not available SNP RefSNP G670A G1377A C834T T/C T31A G57A C/G A79C C435T C3122T T315C G1747A 416Ins/Del G565A C709A G1368A C1528T G1561A C65T C225A A338G 1002R/3R G58C 15705Ins/Del C452T G80A 5UTR T/R 5UTR 3C/G C211T G3809A C3942T C/T A870G Ser31Arg rs1800682 rs2234767 rs763110 rs1800566 rs2273535 rs1047972 rs1862214 rs2072671 rs1048977 rs3775289 N/A N/A rs17215836 rs2290272 rs8187758 rs2242048 rs2242047 rs2242046 rs61637002 rs1060896 rs10868138 N/A N/A N/A N/A rs1051266 N/A N/A N/A rs11568591 rs2277624 rs1805329 rs603965 rs1801270 8 SUPPLEMENTARY REFERENCES: 82. Yu D, Zhang X, Liu J, et al: Characterization of functional excision repair cross-complementation group 1 variants and their association with lung cancer risk and prognosis. Clin Cancer Res 14:2878-86, 2008 83. Camps C, Sarries C, Roig B, et al: Assessment of nucleotide excision repair XPD polymorphisms in the peripheral blood of gemcitabine/cisplatin-treated advanced nonsmall-cell lung cancer patients. Clin Lung Cancer 4:237-41, 2003 84. Wu W, Zhang W, Qiao R, et al: Association of XPD polymorphisms with severe toxicity in non-small cell lung cancer patients in a Chinese population. Clin Cancer Res 15:3889-95, 2009 85. Bepler G, Zheng Z, Gautam A, et al: Ribonucleotide reductase M1 gene promoter activity, polymorphisms, population frequencies, and clinical relevance. Lung Cancer 47:183-92, 2005 86. Feng JF, Wu JZ, Hu SN, et al: Polymorphisms of the ribonucleotide reductase M1 gene and sensitivity to platin-based chemotherapy in non-small cell lung cancer. Lung Cancer, 2009 87. Isla D, Sarries C, Rosell R, et al: Single nucleotide polymorphisms and outcome in docetaxel-cisplatin-treated advanced non-small-cell lung cancer. Ann Oncol 15:1194-203, 2004 88. Kim SO, Jeong JY, Kim MR, et al: Efficacy of gemcitabine in patients with non-small cell lung cancer according to promoter polymorphisms of the ribonucleotide reductase M1 gene. Clin Cancer Res 14:3083-8, 2008 89. Soo RA, Wang LZ, Ng SS, et al: Distribution of gemcitabine pathway genotypes in ethnic Asians and their association with outcome in non-small cell lung cancer patients. Lung Cancer 63:121-7, 2009 90. Feng J, Sun X, Sun N, et al: XPA A23G polymorphism is associated with the elevated response to platinum-based chemotherapy in advanced non-small cell lung cancer. Acta Biochim Biophys Sin (Shanghai) 41:429-35, 2009 91. Hsu HS, Lee IH, Hsu WH, et al: Polymorphism in the hMSH2 gene (gISV126T > C) is a prognostic factor in non-small cell lung cancer. Lung Cancer 58:123-30, 2007 92. Sohn JW, Lee SY, Lee SJ, et al: MDR1 polymorphisms predict the response to etoposide-cisplatin combination chemotherapy in small cell lung cancer. Jpn J Clin Oncol 36:137-41, 2006 93. Muller PJ, Dally H, Klappenecker CN, et al: Polymorphisms in ABCG2, ABCC3 and CNT1 genes and their possible impact on chemotherapy outcome of lung cancer patients. Int J Cancer 124:1669-74, 2009 94. Sasaki H, Endo K, Takada M, et al: EGFR polymorphism of the kinase domain in Japanese lung cancer. J Surg Res 148:260-3, 2008 95. Sasaki H, Okuda K, Takada M, et al: A novel EGFR mutation D1012H and polymorphism at exon 25 in Japanese lung cancer. J Cancer Res Clin Oncol 134:1371-6, 2008 96. Booton R, Ward T, Heighway J, et al: Glutathione-S-transferase P1 isoenzyme polymorphisms, platinum-based chemotherapy, and non-small cell lung cancer. J Thorac Oncol 1:679-83, 2006 97. Lu C, Spitz MR, Zhao H, et al: Association between glutathione S-transferase pi polymorphisms and survival in patients with advanced nonsmall cell lung carcinoma. Cancer 106:441-7, 2006 9 98. Sreeja L, Syamala V, Hariharan S, et al: Glutathione S-transferase M1, T1 and P1 polymorphisms: susceptibility and outcome in lung cancer patients. J Exp Ther Oncol 7:73-85, 2008 99. Yang P, Yokomizo A, Tazelaar HD, et al: Genetic determinants of lung cancer short-term survival: the role of glutathione-related genes. Lung Cancer 35:221-9, 2002 100. Rollin J, Regina S, Vourc'h P, et al: Influence of MMP-2 and MMP-9 promoter polymorphisms on gene expression and clinical outcome of non-small cell lung cancer. Lung Cancer 56:273-80, 2007 101. Matakidou A, El Galta R, Rudd MF, et al: Further observations on the relationship between the FGFR4 Gly388Arg polymorphism and lung cancer prognosis. Br J Cancer 96:1904-7, 2007 102. Spinola M, Leoni V, Pignatiello C, et al: Functional FGFR4 Gly388Arg polymorphism predicts prognosis in lung adenocarcinoma patients. J Clin Oncol 23:7307-11, 2005 103. Ando M, Ando Y, Sekido Y, et al: Genetic polymorphisms of the UDPglucuronosyltransferase 1A7 gene and irinotecan toxicity in Japanese cancer patients. Jpn J Cancer Res 93:591-7, 2002 104. Font A, Sanchez JM, Taron M, et al: Weekly regimen of irinotecan/docetaxel in previously treated non-small cell lung cancer patients and correlation with uridine diphosphate glucuronosyltransferase 1A1 (UGT1A1) polymorphism. Invest New Drugs 21:435-43, 2003 105. Han JY, Lim HS, Shin ES, et al: Influence of the organic anion-transporting polypeptide 1B1 (OATP1B1) polymorphisms on irinotecan-pharmacokinetics and clinical outcome of patients with advanced non-small cell lung cancer. Lung Cancer 59:69-75, 2008 106. Alberola V, Sarries C, Rosell R, et al: Effect of the methylenetetrahydrofolate reductase C677T polymorphism on patients with cisplatin/gemcitabine-treated stage IV nonsmall-cell lung cancer. Clin Lung Cancer 5:360-5, 2004 107. Takehara A, Kawakami K, Ohta N, et al: Prognostic significance of the polymorphisms in thymidylate synthase and methylenetetrahydrofolate reductase gene in lung cancer. Anticancer Res 25:4455-61, 2005 108. Fong KM, Kida Y, Zimmerman PV, et al: MYCL genotypes and loss of heterozygosity in non-small-cell lung cancer. Br J Cancer 74:1975-8, 1996 109. Shih CM, Kuo YY, Wang YC, et al: Association of L-myc polymorphism with lung cancer susceptibility and prognosis in relation to age-selected controls and stratified cases. Lung Cancer 36:125-32, 2002 110. Spinola M, Falvella FS, Galvan A, et al: Ethnic differences in frequencies of gene polymorphisms in the MYCL1 region and modulation of lung cancer patients' survival. Lung Cancer 55:271-7, 2007 111. Tefre T, Borresen AL, Aamdal S, et al: Studies of the L-myc DNA polymorphism and relation to metastasis in Norwegian lung cancer patients. Br J Cancer 61:809-12, 1990 112. Colakogullari M, Ulukaya E, Yilmaztepe Oral A, et al: The involvement of IL-10, IL-6, IFN-gamma, TNF-alpha and TGF-beta gene polymorphisms among Turkish lung cancer patients. Cell Biochem Funct 26:283-90, 2008 113. Kotsinas A, Evangelou K, Sideridou M, et al: The 3' UTR IGF2R-A2/B2 variant is associated with increased tumor growth and advanced stages in non-small cell lung cancer. Cancer Lett 259:177-85, 2008 114. Pine SR, Mechanic LE, Ambs S, et al: Lung cancer survival and functional polymorphisms in MBL2, an innate-immunity gene. J Natl Cancer Inst 99:1401-9, 2007 10 115. Buckingham LE, Coon JS, Morrison LE, et al: The prognostic value of chromosome 7 polysomy in non-small cell lung cancer patients treated with gefitinib. J Thorac Oncol 2:414-22, 2007 116. Manenti G, De Gregorio L, Pilotti S, et al: Association of chromosome 12p genetic polymorphisms with lung adenocarcinoma risk and prognosis. Carcinogenesis 18:1917-20, 1997 117. Manenti G, Galbiati F, Pettinicchio A, et al: A V141L polymorphism of the human LRMP gene is associated with survival of lung cancer patients. Carcinogenesis 27:1386-90, 2006 118. Mitsuta K, Yokoyama A, Kondo K, et al: Polymorphism of the MUC1 mucin gene is associated with susceptibility to lung adenocarcinoma and poor prognosis. Oncol Rep 14:185-9, 2005 119. Park JY, Lee WK, Jung DK, et al: Polymorphisms in the FAS and FASL genes and survival of early stage non-small cell lung cancer. Clin Cancer Res 15:1794-800, 2009 120. Spinola M, Meyer P, Kammerer S, et al: Association of the PDCD5 locus with lung cancer risk and prognosis in smokers. J Clin Oncol 24:1672-8, 2006 121. Gautschi O, Hugli B, Ziegler A, et al: Cyclin D1 (CCND1) A870G gene polymorphism modulates smoking-induced lung cancer risk and response to platinum-based chemotherapy in non-small cell lung cancer (NSCLC) patients. Lung Cancer 51:303-11, 2006 122. Shih CM, Lin PT, Wang HC, et al: Lack of evidence of association of p21WAF1/CIP1 polymorphism with lung cancer susceptibility and prognosis in Taiwan. Jpn J Cancer Res 91:9-15, 2000 11