PRO_408_sm_suppfig6
advertisement

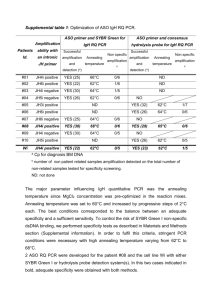
Supplemental Figure 6: Analysis of amplification efficiencies of different primer sets to detected mRNA expression levels Since the nucleotide sequence of optimized and wt DNA differs up to 30 %, a design of uniform primer sets was not possible. Therefore, we first examined the amplification efficiencies of different primer sets using at least 6-log template dilution series to ensure the comparability of the detected mRNA expression levels. Several primer pairs were tested for each target to identify sets for the wt and optimized sequence that showed matching amplification efficiencies and annealing temperatures such that wt and optimized samples could be run on the same plate and results could be directly compared. For the evaluation of the amplification efficiencies and linear ranges of the qPCR assays, standard curves of cDNA were produced and threshold cycles (Ct) were plotted against log10 of template concentrations. Amplification efficiencies were calculated from the slope of the graph using E = 10(1/slope) . The comparability of amplification efficiencies of wt and optimized targets were checked by plotting the differences in Ct values between the targets against the logarithm of the template amount. If the slope of the resulting straight line is <0.1, amplification efficiencies are comparable. The specificity of primer pairs was assessed by melting curve analysis. Results for the amplification efficiencies are shown in Supplemental Figure 6. The chosen primers are listed in Supplemental Table 2. Supplemental Figure 6: Analysis of amplification efficiency of different primer sets. Different primer sets for relative quantification of mRNA levels of CK1 (A) and LCK (B) were tested for their amplification efficiencies by running a standard curve with a 6-7 log serial dilution of the template. Threshold cycles (Ct) were plotted against log10 of the template concentration and the amplification efficiency was calculated as E = 10(1/slope) from the resulting straight line.