Supplemental table 1: Optimization of SYBR Green and
advertisement

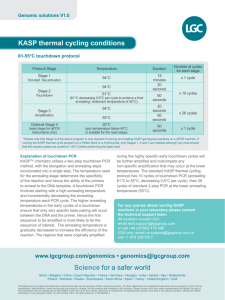
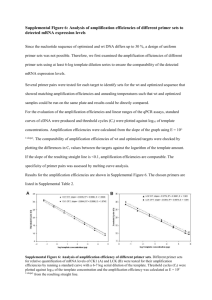
Supplemental table 1: Optimization of ASO IgH RQ PCR. ASO primer and SYBR Green for ASO primer and consensus IgH RQ PCR hydrolysis probe for IgH RQ PCR Amplification Patients ability with Id. Successful an intronic amplification Annealing JH primer and temperature Non specific amplification b detection (a) Successful amplification Annealing and temperature Non specific amplification b detection (a) #01 JH4i positive YES (25) 66°C 0/6 ND #02 JH6i positive YES (22) 62°C 1/6 ND #03 JH4i negative YES (30) 64°C 1/5 ND #04 JH5i negative YES (26) 62°C 0/6 #05 JH3i positive ND YES (32) 62°C 1/7 #06 JH6i positive ND YES (26) 64°C 0/6 #07 JH6i negative YES (25) 64°C 0/6 NO #08 JH4i positive YES (30) 68°C 0/6 YES (29) 65°C 0/6 #09 JH4i negative YES (30) 64°C 0/5 NO #10 JH5i positive YES (26) 62°C 0/5 Wi JH4i positive YES (23) 62°C 1/5 ND YES (22) 62°C 0/5 NO a Cp for diagnosis BM DNA b number of non patient related samples amplification detected on the total number of non-related samples tested for specificity screening. ND: not done The major parameter influencing IgH quantitative PCR was the annealing temperature since MgCl2 concentration was pre-optimized in the reaction mixes. Annealing temperature was set to 60°C and increased by progressive steps of 2°C each. The best conditions corresponded to the balance between an adequate specificity and a sufficient sensitivity. To control the risk of SYBR Green I non-specific dsDNA binding, we performed specificity tests as described in Materials and Methods section (Supplemental information). In order to fulfil this criteria, stringent PCR conditions were necessary with high annealing temperature varying from 62°C to 68°C. 2 ASO RQ PCR were developed for the patient #08 and the cell line Wi with either SYBR Green I or hydrolysis probe detection system(s). In this two cases indicated in bold, adequate specificity were obtained with both methods.