Human SNPs Genotyping by Single Base Extension on
advertisement

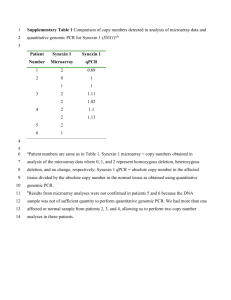
Human SNPs Genotyping by Single Base Extension on PNA Microarray Jae Yang Song, Hyun Gyu Park Department of Chemical and Biomolecualr Engineering Korea Advanced Institute of Science and Technology 373-1 Guseong-dong, Yuseong-gu, Daejeon 305-701 Republic of Korea Abstract: Large scale human genetic studies require technologies for generating millions of genotypes with relative ease but also at a reasonable cost and with high accuracy. We describe a novel system that allows high-throughput genotyping of single nucleotide polymorphisms (SNPs) on PNA microarray by single base extension (SBE) reaction. SBE reaction use bifunctional primers carrying a unique sequence tag in addition to a locus specific sequence. SBE primers are extended with biotin-labeled ddNTPs and stained with streptavidin-R-phycoerythrin after hybridization on PNA microarray. Seven SNPs related with diabetes disease were detected successfully through single-tube-SBE reaction. Genotypes are deduced from the fluorescence signals. Higher degrees of multiplexing would be possible with this method. Key-Words: - PNA, SBE, Multiplex genotyping, Microarray, 1 Introduction experiments such as DNA array, Northern or Southern bolt, FISH, detection of single point mutations and DNA mapping. This paper describes a novel enzyme assisted method that allows efficient high-throughput genotyping of multiplex amplified genome SNPs in a microarray format. The method is based on the single base extension reaction that using chimeric primer with 3 complementarity to the specific SNP loci and 5 complementarity to specific probes, or tags, synthesized on microarray. Genomewide genetic analysis of gene-based single nucleotide polymorphisms (SNPs) may be an efficient paradigm for the discovery of genes important in complex traits in humans (1). Methods to screen and map genetic variability have, for more than two decades, been used on restriction fragment length polymorphism and microsatellite markers. Identification of complex disease genes will require both linkage and association analysis of thousands of individuals. To enable such large-scale polymorphism analysis in human studies, parallel and efficient genotyping methods are critically needed. 2 Materials and Methods A wide variety of technologies have been used to genotype SNPs, including single base extension, 2.1 DNA sample TaqMan assay (2), oligonucleotide ligation assay (3), Sample of known genotype analyzed in our study was molecular beacon (4) and cleavage by a flab provided by voluntary personal. The DNA was endonuclease (5). extracted from whole blood by a standard method. We used PNA microarray in this experiment. Peptide nucleic acid (PNA) is novel DNA mimic in which the 2.2 Primers sugar-phosphate backbone has been replaced with a A pair of PCR primers were designed to have a similar backbone based on amino acid (6). PNA exhibit Tm near 70 C, a G+C content of near 50%, and a sequence-specific binding to DNA and RNA with length of 42 mer and 37 mer with a product size 361 bp. higher affinity and specificity than unmodified DNA. Forward primer had a 5 tail of T7 sequence PNA is resistant to nuclease and protease attack in (TAATACGACTCACTATAGGG) and the reverse serum and cellular extracts and, thus, appear very promising as diagnostic and biomolecular probes, and primer had a 5 tail of T3 sequence possibly as antisense and antigene drugs. For instance, (ATTAACCCTCACTAAA). SBE primers were PNA labeled with biotin, fluorescent dyes or reporter designed as bifunctional primers carrying a unique enzymes are powerful probes in hybridization Table 1. Oligonucleotide sequences for multiplex SBE genotyping for 7 SNPs. * All sequences are written 5 to 3. SNP Allele SNP1 SNP2 SNP3 SNP4 SNP5 SNP6 SNP7 G G G C C C C Zip Code 1 2 9 3 4 7 6 Capture Probe Sequence TGCGGGTAATCG ATCGTGCGACTT GGTAATCGACCT ATCGACTTTGCG CAGCACCTTGCG ATCGGGTATGCG TGCGACCTGGTA SBE Primer Sequence CGATTACCCGCAGCAGCACAACATCCCACAGC AGGTCGCACGATCCCTAACTCATAGGT AGGTCGATTACCAACACCTCAACAA CGCAAGGTCGATACTCCCATGAAGACGCAGAAG CGCAAGGTGCTGCACTGGCCTCAACCAGTCC CGCATACCCGATAGCAGCACAACATCCCACAG TACCAGGTCGCAGATACCACTGGCCTCAACCAGT The microarrays were prepared on standard microscope glass slide. PNA (Peptide nucleic acid) 12 mer oligonucleotide contained a 5 NH2 group spotted on microarray. To reduce the spot variation, each sequence has 9 (33) spots respectively. The biotin labeled SBE reaction products were denatured at 95 C for 5 minutes and snap cooled on ice for 3-5 minutes, then hybridized with 30 l hybridization solution (1X SSPET, 0.01 % Triton X-100) at 37 C for 12 hours. After hybridization, the arrays were rinsed with 6X SSPET (0.005 % Triton X-100) for 10 minutes at room temperature and then stained at 37 C for 2 hours with 30 l staining solution (3 g/ml streptavidin R-phycoerythrin in 1X SSPET 0.01% Triton X-100). 2.4 PCR Amplification 2.7 Array Scanning and Signal Quantiation The amplifications were carried out using 100 ng of DNA, 0.2 mM dNTPs, 100 M each primer, 10 mM Tris HCl, 40 mM KCl, 1.5 mM MgCl2, and 2.5 U Taq DNA polymerase (Bioneer) in 50 l of reaction buffer. After initial activation of the polymerase at 94 C for 5 minutes, the thermocycling parameters were as follows: 35 cycles of 94 C for 30 seconds, 55 C for 30 seconds, 72 C for 1 minutes, and final extension at 72 C for 7 minutes. The microscope glass slides were scanned using the fluorescence scanner GenePix4000B (Axon instrument). 16 bit TIFF images were analyzed using GenePix Pro 3.0 software. sequence tag in addition to a locus-specific sequence (Table1). 2.3 Microarray Design 2.5 Multiplex SBE reaction SBE reactions were carried out in 20 l reactions using 8 l the template, 0.25 M each SBE primer, 2 U Thermosequenase (Amersham), 26 mM Tris-HCl. 6.5 mM MgCl2 25 M biotin-ddGTP, 25 M biotin-ddCTP. Single base extension reactions were carried out on a thermocycler (AppliedBiosystems) with initial polymerase activation at 96 C for 5 minutes, then 40 cycles of 96 C for 30 seconds, 50 C for seconds and 60 C for 2 minutes. After SBE reaction, the products of SBE were purified using QIAquick Nucleotide Removal Kit (Qiagen). 2.6 Hybridization on PNA Tag Array 3 Results and Discussion 3.5 Reaction Principle Figure 1 shows the principle and procedure of the novel single base extension method for genotyping single nucleotide variation on microarray. Double stranded PCR products serves as template for the SBE reaction. Each SBE primer is chimeric with a 5 end complementary to a unique tag synthesized on the array a 3 end complementary to the genomic sequence and terminating one base before a polymorphic SNP site. Thus, each SBE primer is uniquely associated with a specific tag on the array. SBE primers corresponding to multiple markers are added to a single reaction tube and extended in the presence of biotin-labeled ddNTPs. The labeled multiplex SBE reaction products are pooled and hybridized to the tag array. Figure 1. Schematic of SBE genotyping on PNA microarray. Multiplex SBE reactions are performed in single tube; Each SBE primer is chimeric with unique sequence tag. Multiplex SBE reaction analyzed on PNA microarray after hybridization. Probes on microarray has complemetary sequences to each chimeric SBE primer . 3.6 Genotype Discrimination Single base extension involves extension of a primer located adjacent to the position of a SNP, using DNA polymerase in the presence of biotin-labeled ddNTPs. We demonstrate the potential for accurate multiplex genotyping by the single base extension by applying it to three SNPs in single tube reaction. The result shows high fidelity genotype discrimination for three SNPs in one step single base extension reaction.(Figure 2B). We also evaluated our single base extension method with seven SNPs (Figure 2C). These reaction conditions were identical to the optimized conditions applied in this first assay. Because of its accuracy, ease, and potential for high throughput, our novel microarray-based method will greatly benefit various large scale SNP genotyping or mutation screening project. (A) (B) ( C) Figure 2. (A) The design of an PNA microarray is illustrated schematically. Twenty groups of spots have been formed on microslide. Each group has nine spots to reduce the fluorescence intensity variation. The entire PNA microarray contains 20 unique tag sequence of 12 mer probe. (B) Fluorescence image of PNA microarray following hybridization of 3 SNPs SBE primers. (C) Fluorescence image of seven SNPs genotyping by multiplex SBE reactions on PNA microarray. References: [1] Risch, N. & Merikangas, K., Science, 273,1996,pp 1516-1517. [2] Livak, K. J., Marmaro, J. & Todd J. A., Nat. Genet.,9, 1995, pp 341-342. [3] Tobe, V. O., Taylor, S. L. & Nickerson, D. A., Nucleic Acids, 24, 1996, pp 3728-3732. [4] Tyagi, S., Bratu, D. P. & Kramer, F. R., Nat. Biotechnol. 16, 1998, pp 49-53. [5] Mein, C. A., Barratt, B. J., Dunn, M. G., Siemund, T., smith, A. N., Esposito, L., Nutland, S., Stevens, H. E., Wilson, A. J., Phillips, M. S., et al., Genome Res.,10,2000,pp330-343. [6] Peter E. N., Acc. Chem. Res., 32, 1999, pp 624-630.