Title: Copper Homeostasis in Drosophila Melanogaster S2 Cells
advertisement

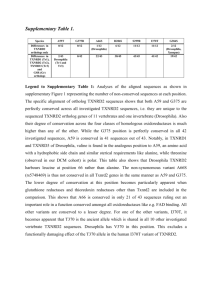
Supplementary material Fig. S1. 1 Fig. S2A. 2 Fig. S2B. 3 G Fig. S3. 4 References Cherbas, L. (2004). General procedures for maintenance of Drosophila cell lines. Indiana University: Drosophila Genomics Resource Center. Russo, C. A., Takezaki, N. and Nei, M. (1995). Molecular phylogeny and divergence times of drosophilid species. Mol Biol Evol 12, 391-404. Rutherford, K., Parkhill, J., Crook, J., Horsnell, T., Rice, P., Rajandream, M. A. and Barrell, B. (2000). Artemis: sequence visualization and annotation. Bioinformatics 16, 944-5. Thompson, J. D., Higgins, D. G. and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22, 4673-80. 5 Supplementary figure captions Fig. S1. Copper-transporting PIB-type ATPase protein sequence alignment. Human ATP7A (NP000043.3), human ATP7B (NP000044.2), mouse ATP7A (NP001103227.1), mouse ATP7B (NP031537.2) and DmATP7 (NP572756.2) were aligned using a default ClustalW protein sequence alignment (Thompson et al., 1994) with Boxshade used to highlight conserved residues. The metal binding domains (GMT/HCxxC), CPC motif and the A (TGE), P (DKTG, TGDN, GDGxND) and N domains (SEHP) are highlighted (box). The di-leucine, PDZ and FAFDNVGYE trafficking motifs are also highlighted (box). Residues phosphorylated in human ATP7A are indicated (). Fig. S2. Drosophila copper-transporting PIB-type ATPase protein sequence alignment and phylogenetic analysis. Putative Drosophila P(IB)-type ATPase orthologues were identified by tBLASTn searches of the genomes available on Flybase (http://flybase.org/) for sequences similar to DmATP7. Single orthologues were found for an additional eleven Drosophila species (simulans, sechellia, yakuba, erecta, ananassae, pseudoobscura, persimilis, willistoni, mojavensis, virilis and grimshawi). Open reading frames were manually re-annotated using Artemis (Rutherford et al., 2000) based on sequence similarity with closely related species. (A) Protein sequences were aligned using ClustalW (Thompson et al., 1994) with Boxshade used to highlight conserved residues. Each orthologous gene contained four exons and intron splice sites are indicated (). The conserved CPC motif, A, P and N domains and four metal binding domains are highlighted (box). The CPC motif could not be identified for D. 6 sechellia due to incomplete sequencing in the region. A putative C-terminal di-leucine motif and Class I PDZ domain (S/TTEL) were conserved in Drosophila orthologues (box), whereas the ATP7B specific FAFDNVGYE motif was not. Two serine residues phosphorylated in ATP7A were also conserved in Drosophila (). (B) Phylogenetic analysis with 100 replicate bootstrapped trees was conducted with the Caenorhabditis elegans ATP7 used as an out group (fpropars, EMBOSS). The relationship between these orthologues closely resembles that of the relationship between the species (Russo et al., 1995). Bioinformatics analysis was conducted using Biomanager 2.0 (Angis, The University of Sydney, NSW, Australia). Fig. S3. DmATP7 does not undergo copper-responsive trafficking in Bm3-c2 cells. The clonal neuronal cell line Dm-BM3-c2, originally isolated from the ventral ganglion of third instar D. melanogaster larvae were purchased from the Drosophila Genomics Resource Center (Bloomington, IN, USA). Cells were maintained in M3 media (SigmaAldrich) supplemented with 10% fetal calf serum (Trace Scientific) and 10 µg/ml insulin (Sigma-Aldrich) at 25C as previously reported (Cherbas, 2004). (A-E) Immunocytochemical detection of DmATP7 in Drosophila BM3-c2 cells exposed to basal media (A-C) or media supplemented with 0.8 mM copper for 2.5 h (D-F). DmATP7 was detected with anti-ATP7B (Green: A, D). Anti-Golgin 97 was used to detect the TGN (Red: B, E) and DAPI was used to detect the nucleus (Blue). Merged images are also shown (C, F). Scale bar = 10 µm. DmATP7 partially co-localized with Golgin 97 at the TGN under both basal and elevated copper conditions. (G) Western blot detection of DmATP7 in BM3-c2 cells exposed to basal media or media 7 supplemented with 0.8 mM copper for 2.5 h. Biotin was used to label cell surface proteins as described in the materials and methods. Biotinylated samples represent protein at the cell surface whereas non-biotinylated samples represent intracellular protein, with total lysate also shown. DmATP7 was detected with anti-ATP7A in the biotinylated sample demonstrating that this protein is present at the PM, under both basal and elevated copper conditions. 8