BSP_Final_Presentation - the Biology Scholars Program Wiki
advertisement

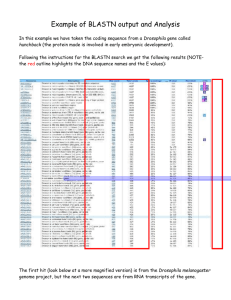
Course Goal Know and use computational biology to predict the evolutionary relatedness and function of proteins Learning Objectives - use computation biology databases, such Blast, SwissPro, and Flybase to identify homologues and predict putative function of a protein Cognitive Level -application -interpretation -evaluation Summative Assessment Upload Final Annotation Report of an unknown Drosophila genomic sequence including all gene features, predicted proteins, and functional homologues Cognitive Level -application -interpretation Formative Assessment You have been assigned the following Drosophila melanogaster protein, SWA1 (swallow). Using the listed databases, completely annotate the gene and confirm the predicted function based on known homologs. Blooms Category: Application, Analysis and Evaluation You have been assigned the following Drosophila melanogaster, SWA1 (swallow). Using Flybase, Genbank, NCBI blast, and SwissPro and Genescan complete the following: a. Retrieve the genomic sequence as a FASTA file (store in Sequence Folder on the Wiki) b. Using NCBI blast perform a tblastx search on your gene sequence. Capture and store on the Wiki. c. Determine the top ten hits based on E value. Report these on your data sheet. d. Determine the first related species not in Drosophila. Report this on your data sheet. e. Using SwissPro analyze the protein sequence of your gene for functional domains, binding motifs, structural features. Report your findings on your data sheet. f. Determine secondary structural features of your protein. g. Using Genescan, predict the genomic structure of your gene. Include all exons, introns and splice sites in the model. Paste link up on the Wiki. In 1-2 pages report the putative function of your protein based on your interpretation of your results from the above modeling programs. Include potential irregularities in your predictions. This would be an in class and then homework writing assignment.