The Tanford β value
advertisement

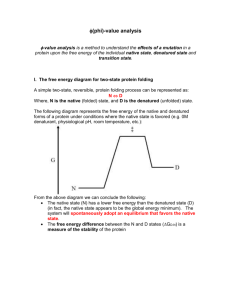
The Tanford β value The transition state of protein folding and unfolding is an intermediate structure between the denatured and native states. Thus, the transition state is stabilized by denaturant with respect to the native structure but destabilized relative to the denatured state. Accordingly, denaturant lowers the activation energy of denaturation and raises the activation energy of folding. If each group, i, in the protein has a free energy of transfer of δgi cal/mol from water to 1-M solution of denaturant and each group becomes exposed by only a factor αi on denaturation, then the effective free energy change for the group on denaturation is αiδgi. This leads to an m value for unfolding of mD N i g i . Then the transition state for unfolding is: i m N i g i i where αi‡ is the fractional degree of exposure in the transition state for denaturation. The Tanford β value, βT, is a measure of the average degree of exposure in the transition state relative to that of the denatured state from the native state. It is a useful index of the compactness of the transition state. /1/ m T N mD N g g i i i i . i i Ф-values When measuring kinetic and thermodynamic effects of mutations in various parts of the protein, the results are typically presented as Ф-values, which represent the change in stability of the transition state accompanying mutation of a residue relative to the effect of the same mutation in the native state. Suppose that a mutation destabilizes the folded structure by ΔΔGN-D units of energy, measured relative to the unfolded state. Then, if the free energy of transition state, measured relative to the unfolded state, changes by ΔΔG‡-D, Ф is defined by /1/ F G D G N D A value Ф ≈ 1 means that the energy of the transition state is perturbed by the mutation by the same amount as the native state is perturbed i.e. the residue is found in a highly structured region of the transition state and makes interactions that contribute equally to the stability of the transition state and the native state. Ф ≈ 0 means that the energy of transition state is perturbed on mutation by the same amount as the denatured state is perturbed i.e. the residue forms very few, if any, interactions with other residues in the transition state. Contact order In the native state there is a correlation between the rate constants for folding of single domain proteins and the average sequence separation between contacting residues. The average sequence distance between all pairs of contacting residues normalized by the total sequence length is relative contact order, CO: /2/ 1 CO LN N S i, j where N is the total number of contacts, ΔSi,j is the sequence separation, in residues, between contacting residues i and j, and L is the total number of residues in protein. There is a very strong correlation between the relative contact order of a protein and the rate at which it folds: proteins that have primarily local contacts (i.e., have a low contact order) tend to fold more rapidly than those that have more non-local interactions (i.e., have a high contact order). A week, but statistically significant, correlation is observed between native state CO and folding transition state placement, θm. Proteins characterized by large contact orders tend to exhibit more well-ordered transition states, presumably because more of the polypeptide must be ordered to form the requisite number of favourable contacts./2/ References: 1. Fersht, A.; Structure and Mechanism in Protein Science; W.H. Freeman and Company, 1999, New York. 2. Plaxco, K.W., Simons, K.T., Baker, D.; Contact Order, Transition State Placement and the Refolding Rates of Single Domain Proteins; J. Mol. Biol. 277, 985-994 (1998). 3. Dobson, C.M.; Experimental investigation of protein folding and misfolding, Methods 34, 414 (2004). 4. Lindorff-Larsen, K., Vendruscolo, M., Paci, E., Dobson, C. ; Transition states for protein folding have native topologies despite high structural variability, Nature Structural & Molecular Biology, 11, (2004).